Probe CUST_25987_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25987_PI426222305 | JHI_St_60k_v1 | DMT400061791 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
All Microarray Probes Designed to Gene DMG402024041
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25835_PI426222305 | JHI_St_60k_v1 | DMT400061787 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25742_PI426222305 | JHI_St_60k_v1 | DMT400061790 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25951_PI426222305 | JHI_St_60k_v1 | DMT400061789 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25987_PI426222305 | JHI_St_60k_v1 | DMT400061791 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
Microarray Signals from CUST_25987_PI426222305
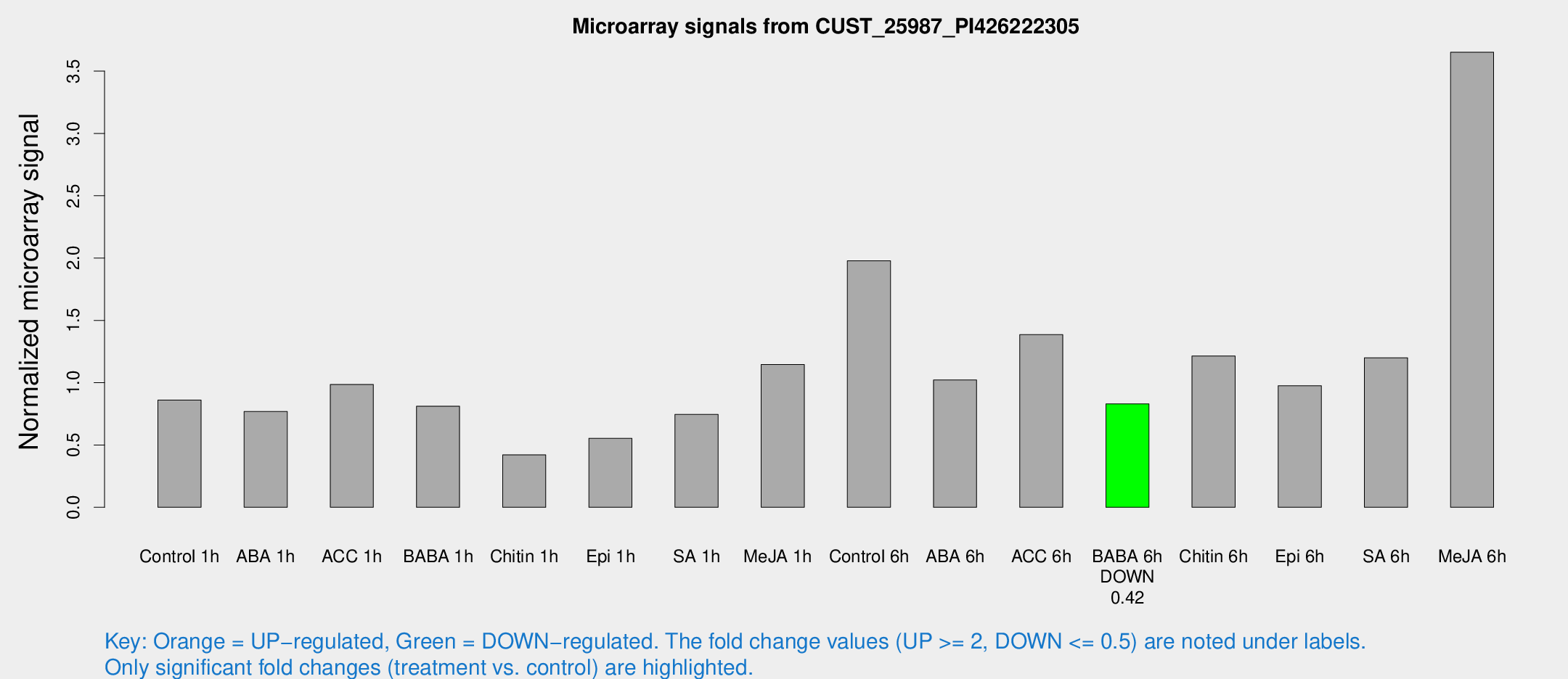
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 83.9706 | 7.00124 | 0.86084 | 0.0618265 |
| ABA 1h | 68.0674 | 12.2424 | 0.768378 | 0.124347 |
| ACC 1h | 100.745 | 16.2801 | 0.985673 | 0.107347 |
| BABA 1h | 77.4894 | 12.1325 | 0.811172 | 0.216982 |
| Chitin 1h | 37.7583 | 6.22225 | 0.420533 | 0.0569593 |
| Epi 1h | 47.2144 | 6.05246 | 0.553424 | 0.0625836 |
| SA 1h | 92.3274 | 38.293 | 0.744995 | 0.506878 |
| Me-JA 1h | 92.0942 | 11.3007 | 1.14613 | 0.207175 |
| Control 6h | 200.644 | 42.7764 | 1.9784 | 0.268821 |
| ABA 6h | 105.865 | 8.63763 | 1.02142 | 0.0701092 |
| ACC 6h | 157.439 | 23.5876 | 1.38531 | 0.318708 |
| BABA 6h | 91.087 | 9.79363 | 0.829572 | 0.0618088 |
| Chitin 6h | 126.143 | 11.1412 | 1.21424 | 0.111706 |
| Epi 6h | 117.272 | 31.5214 | 0.975196 | 0.441254 |
| SA 6h | 119.131 | 22.2802 | 1.20017 | 0.124604 |
| Me-JA 6h | 354.052 | 24.6145 | 3.65183 | 0.386692 |
Source Transcript PGSC0003DMT400061791 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G06350.1 | +2 | 5e-50 | 187 | 116/201 (58%) | dehydroquinate dehydratase, putative / shikimate dehydrogenase, putative | chr3:1924536-1927701 REVERSE LENGTH=603 |
