Probe CUST_2446_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2446_PI426222305 | JHI_St_60k_v1 | DMT400028819 | TGAAAGATGAAGTTGAAAATCTTGCCAGCACGATTCTCCCTCCAAATCTATAAATTGGCG |
All Microarray Probes Designed to Gene DMG402011095
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2070_PI426222305 | JHI_St_60k_v1 | DMT400028820 | TGAAAGATGAACTTGAAAATCTTGCCAGCACGATTCTCCCTCCAAATCTATAAATTGGCG |
| CUST_2446_PI426222305 | JHI_St_60k_v1 | DMT400028819 | TGAAAGATGAAGTTGAAAATCTTGCCAGCACGATTCTCCCTCCAAATCTATAAATTGGCG |
| CUST_2336_PI426222305 | JHI_St_60k_v1 | DMT400028816 | ATGGTTCAAGTTCCCGGGCATGTCTTGTTATTGGGTGCATCTCTACAAGTGAAATTGACG |
Microarray Signals from CUST_2446_PI426222305
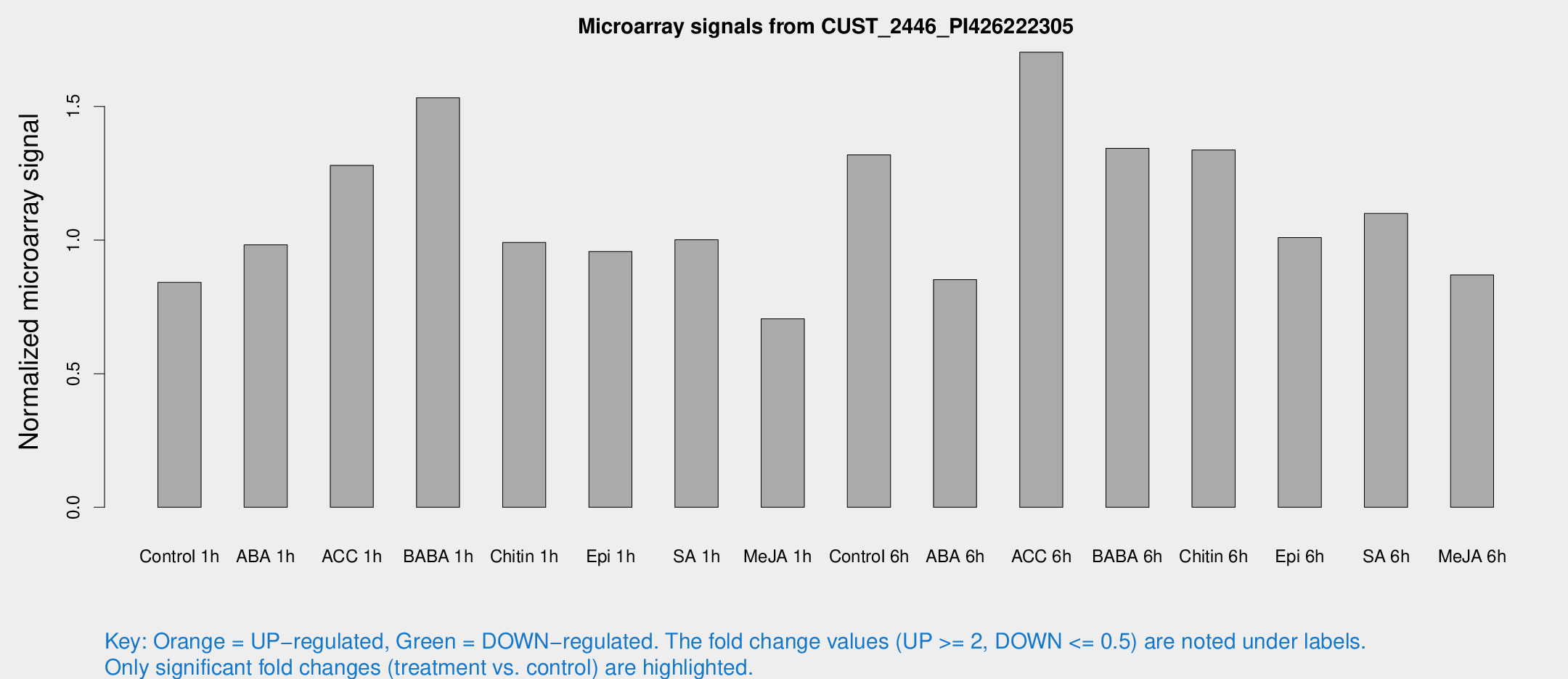
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.4543 | 3.63684 | 0.841561 | 0.16779 |
| ABA 1h | 22.9661 | 3.55592 | 0.982035 | 0.153823 |
| ACC 1h | 40.896 | 17.5483 | 1.27936 | 0.72996 |
| BABA 1h | 40.9918 | 10.0899 | 1.53253 | 0.329132 |
| Chitin 1h | 24.0876 | 4.11921 | 0.991289 | 0.217699 |
| Epi 1h | 22.6087 | 4.18615 | 0.957282 | 0.189189 |
| SA 1h | 27.4342 | 3.71493 | 1.00149 | 0.138354 |
| Me-JA 1h | 15.4076 | 3.64103 | 0.705522 | 0.175666 |
| Control 6h | 35.5083 | 5.13452 | 1.31904 | 0.250489 |
| ABA 6h | 23.885 | 4.03686 | 0.85219 | 0.144733 |
| ACC 6h | 62.9371 | 28.2393 | 1.70301 | 0.951109 |
| BABA 6h | 39.5322 | 4.69883 | 1.34317 | 0.159394 |
| Chitin 6h | 39.0721 | 7.9474 | 1.33717 | 0.316358 |
| Epi 6h | 30.9817 | 5.76526 | 1.00957 | 0.158901 |
| SA 6h | 36.3441 | 14.1463 | 1.09944 | 0.971887 |
| Me-JA 6h | 23.069 | 3.94274 | 0.869463 | 0.151482 |
Source Transcript PGSC0003DMT400028819 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G58770.1 | +2 | 8e-98 | 299 | 145/246 (59%) | Undecaprenyl pyrophosphate synthetase family protein | chr5:23734396-23735495 FORWARD LENGTH=310 |
