Probe CUST_23220_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23220_PI426222305 | JHI_St_60k_v1 | DMT400026660 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
All Microarray Probes Designed to Gene DMG400010302
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23141_PI426222305 | JHI_St_60k_v1 | DMT400026662 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23220_PI426222305 | JHI_St_60k_v1 | DMT400026660 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23183_PI426222305 | JHI_St_60k_v1 | DMT400026663 | GAGTTCAGTAAGCACTCTTACCTTACTCTGCAGGAAGAGGTATTGAAACAAGTGCAGGAC |
| CUST_23185_PI426222305 | JHI_St_60k_v1 | DMT400026661 | GGTAAGGCTTAAGTAGCTCGTATTATACCACTTGGTTTTATAAGTAAACTCCTTAGGATA |
Microarray Signals from CUST_23220_PI426222305
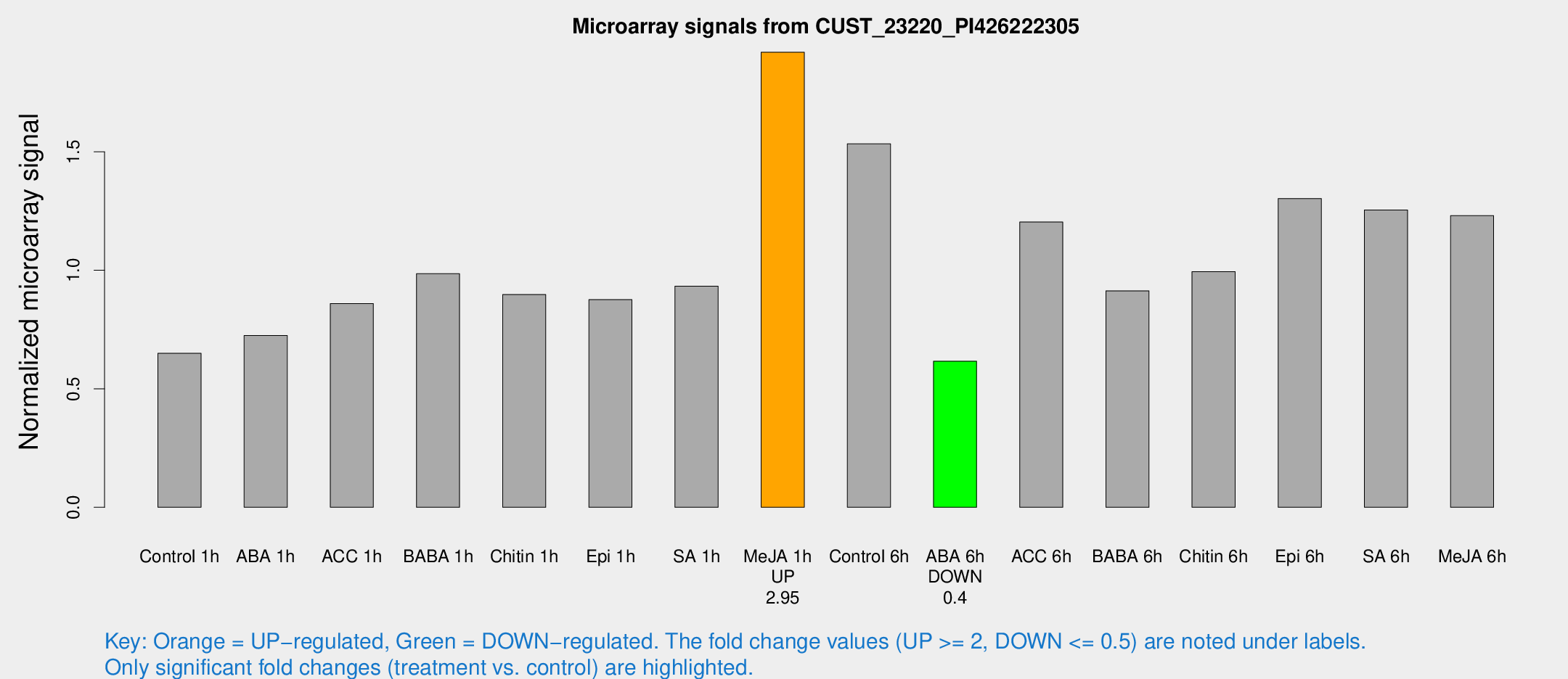
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 79.4246 | 5.64695 | 0.649803 | 0.0462743 |
| ABA 1h | 79.7018 | 10.9476 | 0.724859 | 0.157979 |
| ACC 1h | 109.794 | 15.8493 | 0.859273 | 0.144041 |
| BABA 1h | 116.771 | 10.3783 | 0.985481 | 0.0643806 |
| Chitin 1h | 105.331 | 26.1602 | 0.897713 | 0.312929 |
| Epi 1h | 92.6794 | 6.30378 | 0.87615 | 0.0676614 |
| SA 1h | 122.378 | 27.578 | 0.93313 | 0.208823 |
| Me-JA 1h | 192.654 | 17.6713 | 1.92002 | 0.115955 |
| Control 6h | 196.465 | 39.6649 | 1.53352 | 0.218026 |
| ABA 6h | 80.0187 | 5.82705 | 0.616528 | 0.0448874 |
| ACC 6h | 169.811 | 15.3901 | 1.20366 | 0.142661 |
| BABA 6h | 125.026 | 8.18904 | 0.913405 | 0.0597413 |
| Chitin 6h | 129.162 | 8.34456 | 0.99456 | 0.0642727 |
| Epi 6h | 185.799 | 32.8612 | 1.30275 | 0.175793 |
| SA 6h | 161.203 | 42.1024 | 1.25465 | 0.233731 |
| Me-JA 6h | 150.76 | 13.99 | 1.23048 | 0.0764583 |
Source Transcript PGSC0003DMT400026660 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G44070.1 | +3 | 8e-83 | 276 | 128/157 (82%) | phytochelatin synthase 1 (PCS1) | chr5:17734876-17737672 FORWARD LENGTH=485 |
