Probe CUST_23185_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23185_PI426222305 | JHI_St_60k_v1 | DMT400026661 | GGTAAGGCTTAAGTAGCTCGTATTATACCACTTGGTTTTATAAGTAAACTCCTTAGGATA |
All Microarray Probes Designed to Gene DMG400010302
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23141_PI426222305 | JHI_St_60k_v1 | DMT400026662 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23220_PI426222305 | JHI_St_60k_v1 | DMT400026660 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23183_PI426222305 | JHI_St_60k_v1 | DMT400026663 | GAGTTCAGTAAGCACTCTTACCTTACTCTGCAGGAAGAGGTATTGAAACAAGTGCAGGAC |
| CUST_23185_PI426222305 | JHI_St_60k_v1 | DMT400026661 | GGTAAGGCTTAAGTAGCTCGTATTATACCACTTGGTTTTATAAGTAAACTCCTTAGGATA |
Microarray Signals from CUST_23185_PI426222305
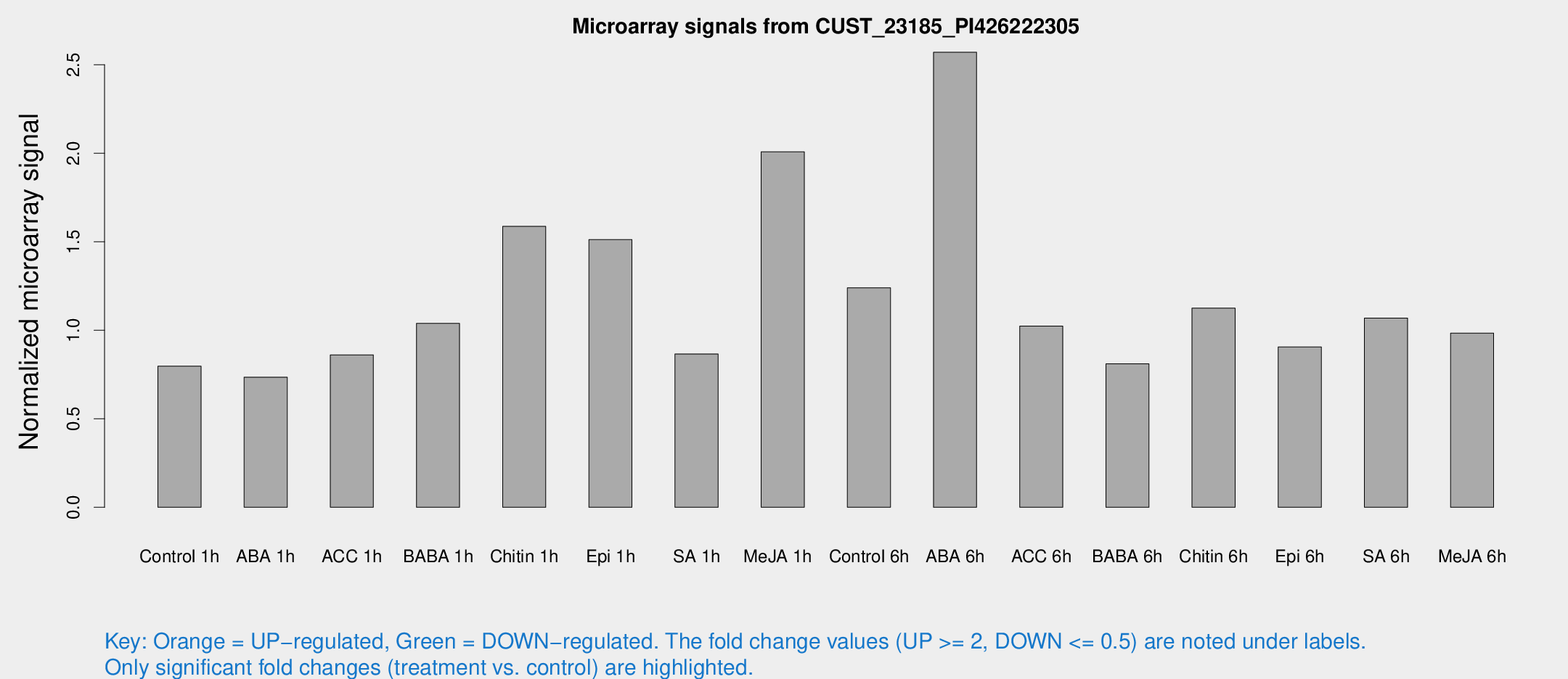
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.66756 | 3.06719 | 0.797368 | 0.349416 |
| ABA 1h | 6.06769 | 2.94531 | 0.73525 | 0.372253 |
| ACC 1h | 8.75621 | 3.40027 | 0.861096 | 0.393077 |
| BABA 1h | 10.0657 | 3.24416 | 1.03889 | 0.403115 |
| Chitin 1h | 14.9378 | 4.54963 | 1.58703 | 0.886589 |
| Epi 1h | 13.8574 | 4.40327 | 1.51199 | 0.771915 |
| SA 1h | 9.98938 | 4.69512 | 0.866274 | 0.433687 |
| Me-JA 1h | 16.031 | 4.13368 | 2.00781 | 0.455343 |
| Control 6h | 13.4008 | 5.2416 | 1.23991 | 0.444561 |
| ABA 6h | 170.925 | 165.325 | 2.5705 | 34.5522 |
| ACC 6h | 10.9792 | 3.90953 | 1.0237 | 0.394643 |
| BABA 6h | 8.63462 | 3.57553 | 0.810283 | 0.357153 |
| Chitin 6h | 12.256 | 3.8877 | 1.12515 | 0.417642 |
| Epi 6h | 9.54463 | 3.69084 | 0.905576 | 0.372159 |
| SA 6h | 10.0894 | 3.37894 | 1.06907 | 0.391911 |
| Me-JA 6h | 10.0022 | 3.25025 | 0.983966 | 0.393087 |
Source Transcript PGSC0003DMT400026661 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G44070.1 | +3 | 1e-125 | 377 | 174/220 (79%) | phytochelatin synthase 1 (PCS1) | chr5:17734876-17737672 FORWARD LENGTH=485 |
