Probe CUST_23141_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23141_PI426222305 | JHI_St_60k_v1 | DMT400026662 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
All Microarray Probes Designed to Gene DMG400010302
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23141_PI426222305 | JHI_St_60k_v1 | DMT400026662 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23220_PI426222305 | JHI_St_60k_v1 | DMT400026660 | TGGAAATTCACGCGAGCACTTGGAAAGACCTTCATTCCTATAATAGAGCTTTCTACCCCC |
| CUST_23183_PI426222305 | JHI_St_60k_v1 | DMT400026663 | GAGTTCAGTAAGCACTCTTACCTTACTCTGCAGGAAGAGGTATTGAAACAAGTGCAGGAC |
| CUST_23185_PI426222305 | JHI_St_60k_v1 | DMT400026661 | GGTAAGGCTTAAGTAGCTCGTATTATACCACTTGGTTTTATAAGTAAACTCCTTAGGATA |
Microarray Signals from CUST_23141_PI426222305
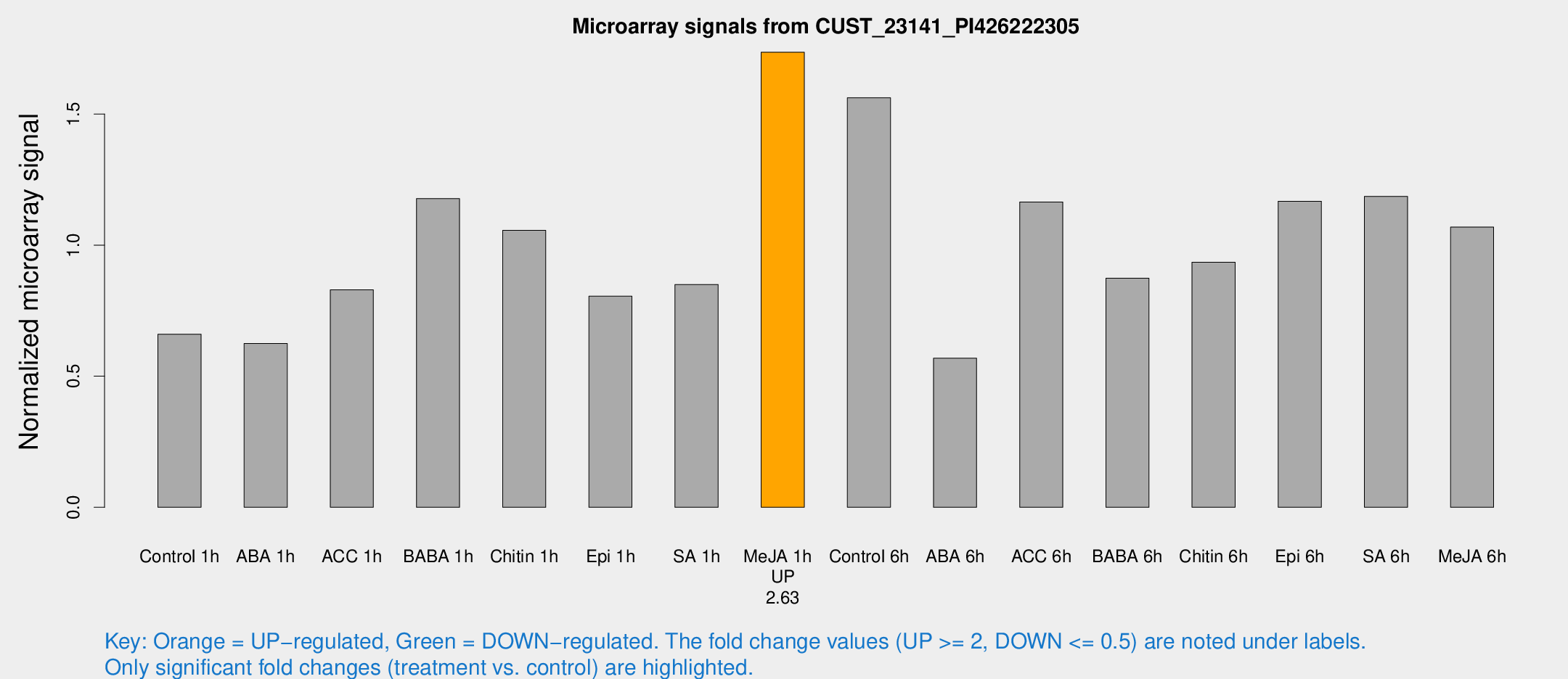
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 90.4705 | 5.99397 | 0.660447 | 0.0437647 |
| ABA 1h | 78.7572 | 16.282 | 0.62457 | 0.174317 |
| ACC 1h | 117.178 | 7.74547 | 0.829451 | 0.11147 |
| BABA 1h | 155.634 | 9.52087 | 1.17732 | 0.172666 |
| Chitin 1h | 136.708 | 30.4284 | 1.05624 | 0.301693 |
| Epi 1h | 96.0115 | 7.36819 | 0.805105 | 0.0847011 |
| SA 1h | 129.924 | 38.0737 | 0.849948 | 0.285278 |
| Me-JA 1h | 196.589 | 21.5619 | 1.73577 | 0.128421 |
| Control 6h | 223.27 | 42.5864 | 1.56196 | 0.200778 |
| ABA 6h | 85.4178 | 14.035 | 0.568785 | 0.0776772 |
| ACC 6h | 184.151 | 14.7355 | 1.16451 | 0.0975065 |
| BABA 6h | 134.08 | 8.46377 | 0.873595 | 0.0551585 |
| Chitin 6h | 136.361 | 8.56386 | 0.934977 | 0.0586939 |
| Epi 6h | 183.633 | 23.8537 | 1.16751 | 0.0779736 |
| SA 6h | 163.755 | 23.4926 | 1.18606 | 0.152021 |
| Me-JA 6h | 148.019 | 17.5604 | 1.06879 | 0.0688636 |
Source Transcript PGSC0003DMT400026662 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G44070.1 | +3 | 0.0 | 568 | 295/474 (62%) | phytochelatin synthase 1 (PCS1) | chr5:17734876-17737672 FORWARD LENGTH=485 |
