Probe CUST_22793_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22793_PI426222305 | JHI_St_60k_v1 | DMT400078022 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
All Microarray Probes Designed to Gene DMG400030345
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22793_PI426222305 | JHI_St_60k_v1 | DMT400078022 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22646_PI426222305 | JHI_St_60k_v1 | DMT400078024 | TATATGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAA |
| CUST_22692_PI426222305 | JHI_St_60k_v1 | DMT400078021 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22582_PI426222305 | JHI_St_60k_v1 | DMT400078020 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
Microarray Signals from CUST_22793_PI426222305
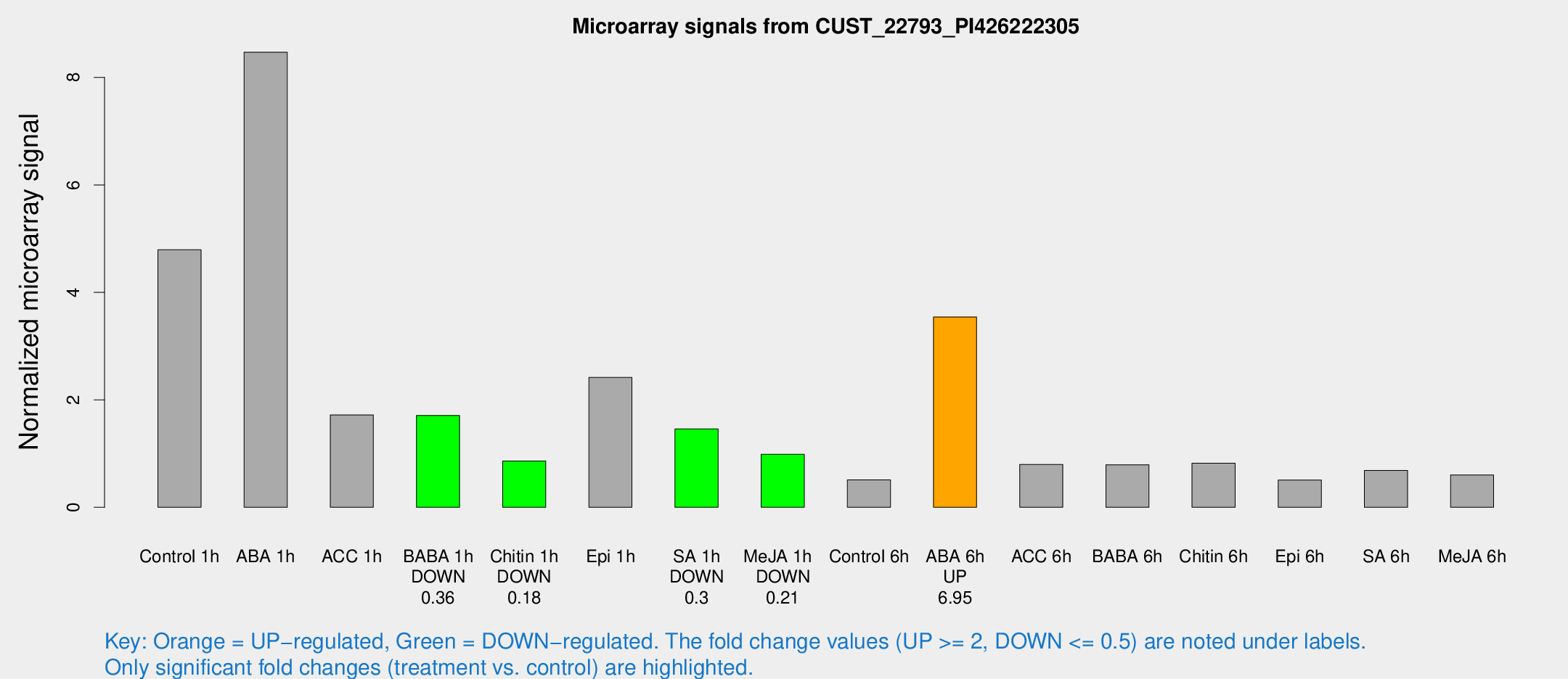
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 909.249 | 61.2413 | 4.7929 | 0.628189 |
| ABA 1h | 1461.19 | 254.078 | 8.46883 | 1.32259 |
| ACC 1h | 449.197 | 207.118 | 1.71831 | 1.12944 |
| BABA 1h | 334.872 | 88.2051 | 1.70902 | 0.327075 |
| Chitin 1h | 150.62 | 24.8392 | 0.863021 | 0.126269 |
| Epi 1h | 415.886 | 98.7609 | 2.41629 | 0.574281 |
| SA 1h | 299.494 | 67.1766 | 1.45941 | 0.313041 |
| Me-JA 1h | 153.081 | 13.0888 | 0.986146 | 0.0602455 |
| Control 6h | 97.0637 | 7.64728 | 0.50973 | 0.0336019 |
| ABA 6h | 742.915 | 141.875 | 3.54284 | 0.620637 |
| ACC 6h | 179.109 | 33.2206 | 0.797157 | 0.201593 |
| BABA 6h | 167.823 | 10.296 | 0.792174 | 0.0484908 |
| Chitin 6h | 169.991 | 26.6754 | 0.822116 | 0.147359 |
| Epi 6h | 108.303 | 7.21774 | 0.506622 | 0.039052 |
| SA 6h | 137.362 | 32.0336 | 0.686496 | 0.109891 |
| Me-JA 6h | 116.174 | 16.9844 | 0.603148 | 0.12298 |
Source Transcript PGSC0003DMT400078022 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07350.1 | +3 | 4e-38 | 145 | 83/109 (76%) | RNA-binding (RRM/RBD/RNP motifs) family protein | chr1:2257732-2260101 REVERSE LENGTH=382 |
