Probe CUST_22646_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22646_PI426222305 | JHI_St_60k_v1 | DMT400078024 | TATATGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAA |
All Microarray Probes Designed to Gene DMG400030345
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22793_PI426222305 | JHI_St_60k_v1 | DMT400078022 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22646_PI426222305 | JHI_St_60k_v1 | DMT400078024 | TATATGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAA |
| CUST_22692_PI426222305 | JHI_St_60k_v1 | DMT400078021 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22582_PI426222305 | JHI_St_60k_v1 | DMT400078020 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
Microarray Signals from CUST_22646_PI426222305
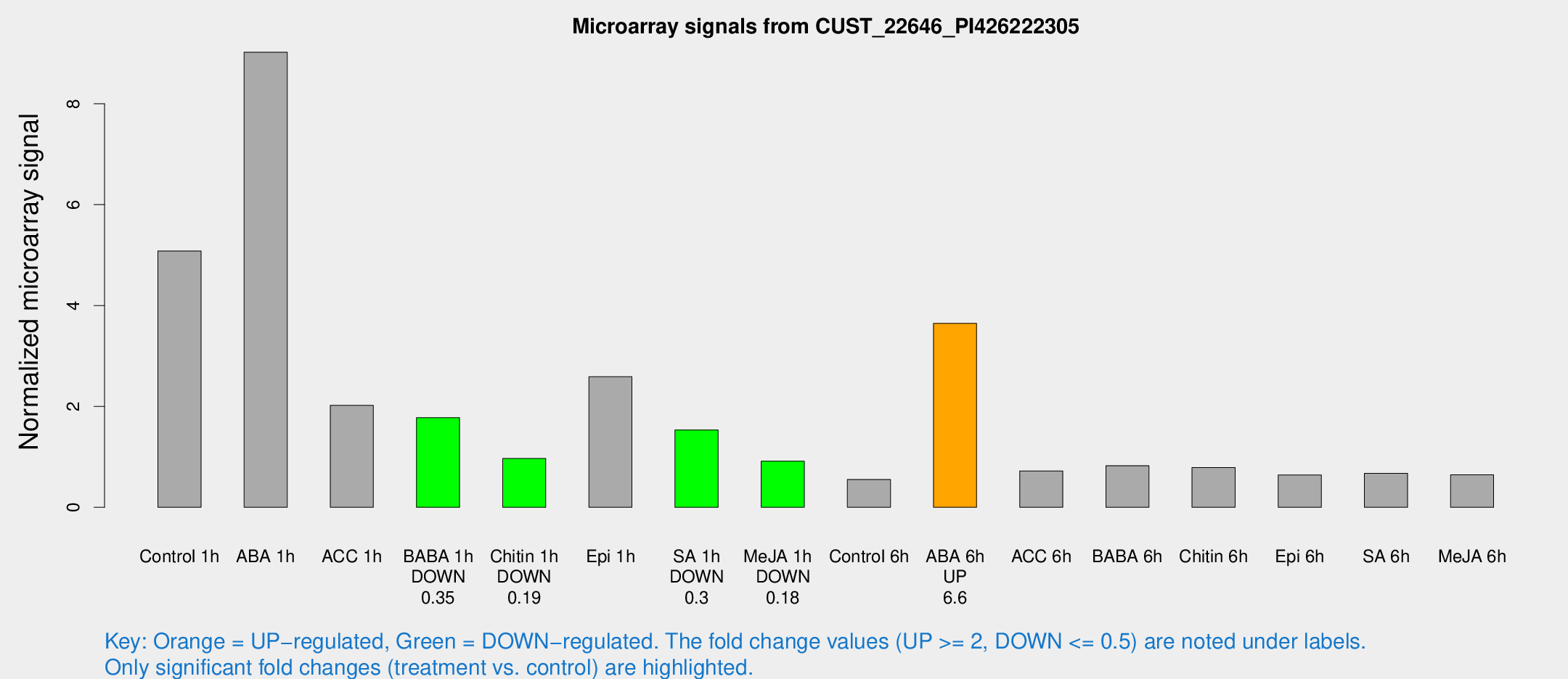
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 854.362 | 57.0827 | 5.07928 | 0.600081 |
| ABA 1h | 1398.52 | 280.076 | 9.02111 | 1.68246 |
| ACC 1h | 427.289 | 177.982 | 2.02129 | 0.943503 |
| BABA 1h | 307.525 | 77.7089 | 1.7756 | 0.323435 |
| Chitin 1h | 152.459 | 34.226 | 0.967103 | 0.141611 |
| Epi 1h | 400.334 | 107.46 | 2.58795 | 0.702637 |
| SA 1h | 278.398 | 61.4362 | 1.5336 | 0.320495 |
| Me-JA 1h | 126.071 | 11.2505 | 0.914947 | 0.0736876 |
| Control 6h | 94.7775 | 13.5948 | 0.552483 | 0.0445302 |
| ABA 6h | 676.753 | 126.674 | 3.64588 | 0.640123 |
| ACC 6h | 151.84 | 48.2809 | 0.720775 | 0.308595 |
| BABA 6h | 156.366 | 15.9144 | 0.824938 | 0.0611872 |
| Chitin 6h | 143.87 | 19.9423 | 0.789935 | 0.125803 |
| Epi 6h | 121.435 | 9.1969 | 0.639518 | 0.0421141 |
| SA 6h | 113.453 | 14.6379 | 0.672739 | 0.0446702 |
| Me-JA 6h | 109.023 | 12.8006 | 0.644722 | 0.0766109 |
Source Transcript PGSC0003DMT400078024 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07350.1 | +3 | 3e-16 | 81 | 38/52 (73%) | RNA-binding (RRM/RBD/RNP motifs) family protein | chr1:2257732-2260101 REVERSE LENGTH=382 |
