Probe CUST_22692_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22692_PI426222305 | JHI_St_60k_v1 | DMT400078021 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
All Microarray Probes Designed to Gene DMG400030345
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22793_PI426222305 | JHI_St_60k_v1 | DMT400078022 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22646_PI426222305 | JHI_St_60k_v1 | DMT400078024 | TATATGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAA |
| CUST_22692_PI426222305 | JHI_St_60k_v1 | DMT400078021 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
| CUST_22582_PI426222305 | JHI_St_60k_v1 | DMT400078020 | TGTTCATGTGGGCGTTCTCCTCGCTGCTAATGGCAGAGAGTATGTTCTTGGAAGAATATA |
Microarray Signals from CUST_22692_PI426222305
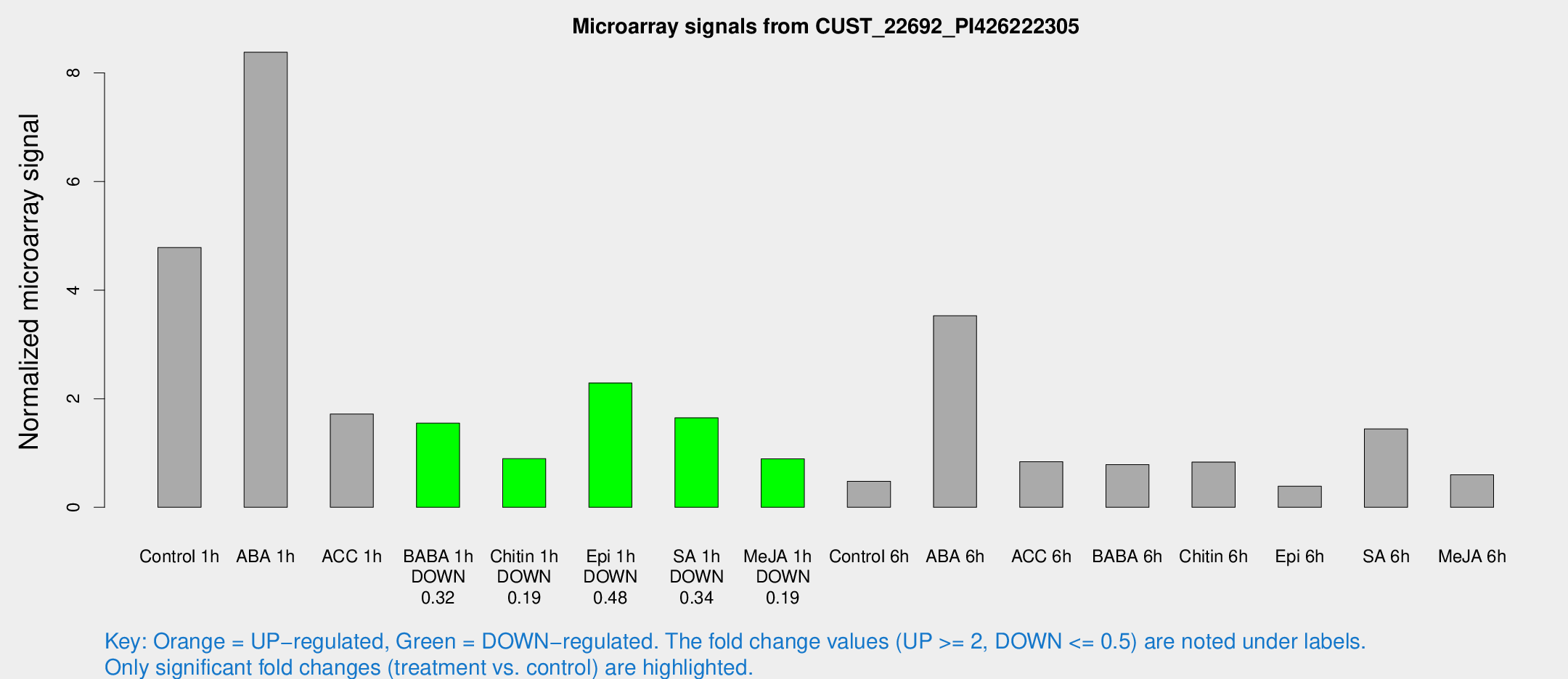
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 861.201 | 49.9831 | 4.78621 | 0.474875 |
| ABA 1h | 1397.38 | 291.943 | 8.38241 | 1.65333 |
| ACC 1h | 415.294 | 180.342 | 1.71859 | 1.04428 |
| BABA 1h | 297.513 | 90.8546 | 1.5506 | 0.384117 |
| Chitin 1h | 149.049 | 25.9058 | 0.896336 | 0.132029 |
| Epi 1h | 366.795 | 66.9691 | 2.29019 | 0.423226 |
| SA 1h | 313.615 | 52.1729 | 1.64914 | 0.234883 |
| Me-JA 1h | 134.671 | 21.8305 | 0.89335 | 0.0802025 |
| Control 6h | 92.0343 | 23.9384 | 0.478215 | 0.0985504 |
| ABA 6h | 699.028 | 124.605 | 3.52942 | 0.555349 |
| ACC 6h | 177.803 | 30.5729 | 0.840118 | 0.22674 |
| BABA 6h | 163.02 | 27.2634 | 0.787371 | 0.109091 |
| Chitin 6h | 159.881 | 10.4409 | 0.833387 | 0.0524323 |
| Epi 6h | 86.7639 | 23.3445 | 0.38949 | 0.144756 |
| SA 6h | 483.996 | 360.306 | 1.4445 | 1.61087 |
| Me-JA 6h | 111.558 | 21.6358 | 0.598221 | 0.134523 |
Source Transcript PGSC0003DMT400078021 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07350.1 | +3 | 5e-38 | 145 | 83/109 (76%) | RNA-binding (RRM/RBD/RNP motifs) family protein | chr1:2257732-2260101 REVERSE LENGTH=382 |
