Probe CUST_18847_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18847_PI426222305 | JHI_St_60k_v1 | DMT400001137 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
All Microarray Probes Designed to Gene DMG401000428
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18800_PI426222305 | JHI_St_60k_v1 | DMT400001138 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18847_PI426222305 | JHI_St_60k_v1 | DMT400001137 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18686_PI426222305 | JHI_St_60k_v1 | DMT400001140 | CGCCAAATATTGGCATAGTAAAGGTATTCTTGGTAGTCCTTTAATCAAATCTCATCTAAG |
| CUST_18731_PI426222305 | JHI_St_60k_v1 | DMT400001134 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18748_PI426222305 | JHI_St_60k_v1 | DMT400001136 | ATAGTTTTGCACTCAGTACAGTTTTTGCTATTGTAGGGTGGTGCTGCTGTCTGGTGCATG |
Microarray Signals from CUST_18847_PI426222305
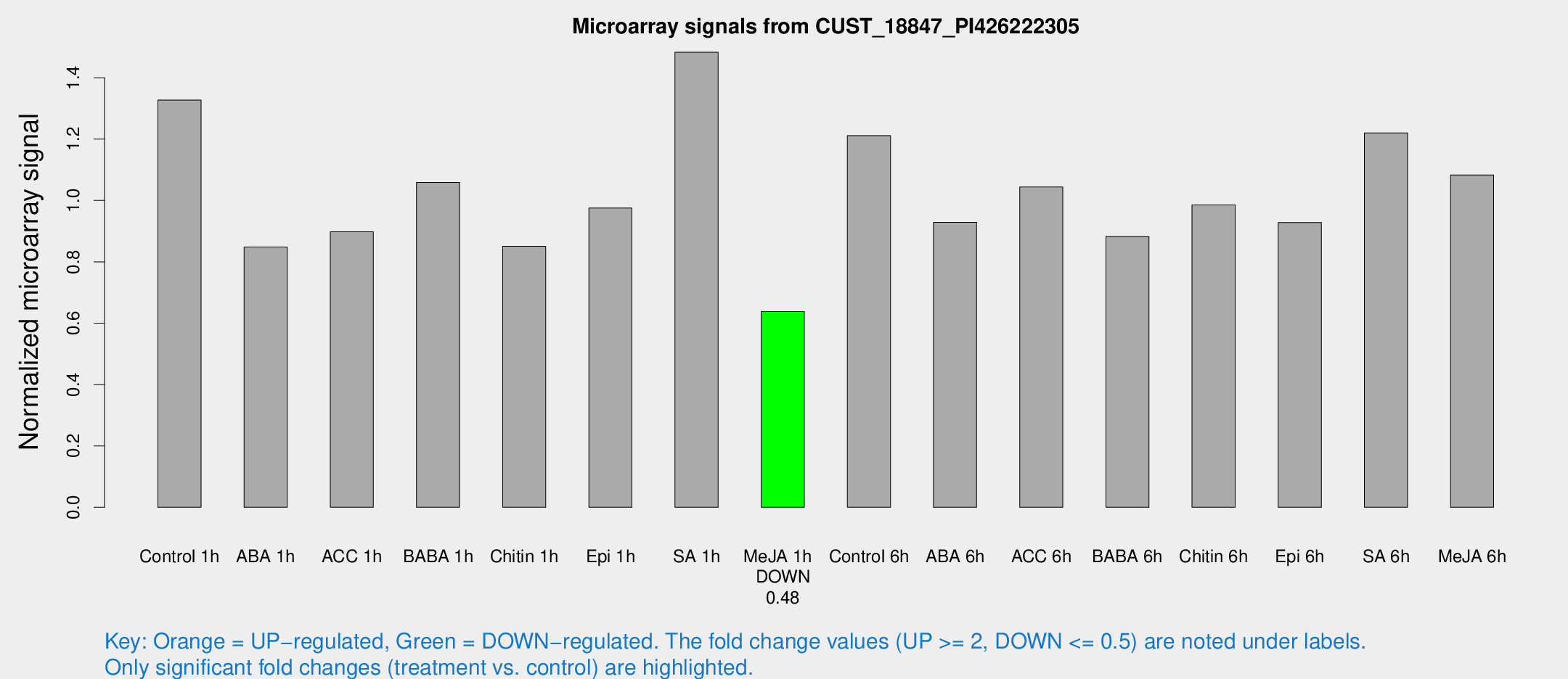
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2371.84 | 213.225 | 1.32727 | 0.0766551 |
| ABA 1h | 1333.38 | 77.2426 | 0.848261 | 0.0527015 |
| ACC 1h | 1821.83 | 513.245 | 0.89797 | 0.262615 |
| BABA 1h | 1875.28 | 334.613 | 1.05859 | 0.100411 |
| Chitin 1h | 1363.18 | 96.602 | 0.850923 | 0.0491773 |
| Epi 1h | 1519.68 | 179.546 | 0.975464 | 0.112807 |
| SA 1h | 2735.76 | 306.041 | 1.48305 | 0.134652 |
| Me-JA 1h | 930.067 | 80.3166 | 0.637727 | 0.0369009 |
| Control 6h | 2233.41 | 413.059 | 1.21131 | 0.149078 |
| ABA 6h | 1784.4 | 246.875 | 0.928527 | 0.0794232 |
| ACC 6h | 2137.21 | 174.728 | 1.04364 | 0.134589 |
| BABA 6h | 1780.94 | 219.678 | 0.882534 | 0.0859632 |
| Chitin 6h | 1865.68 | 109.261 | 0.985388 | 0.0569297 |
| Epi 6h | 1859.31 | 107.642 | 0.928363 | 0.0536395 |
| SA 6h | 2306.64 | 571.002 | 1.22003 | 0.213554 |
| Me-JA 6h | 1969.79 | 348.631 | 1.08318 | 0.102911 |
Source Transcript PGSC0003DMT400001137 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G22850.1 | +3 | 0.0 | 662 | 327/474 (69%) | Eukaryotic aspartyl protease family protein | chr5:7633717-7636298 REVERSE LENGTH=493 |
