Probe CUST_18731_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18731_PI426222305 | JHI_St_60k_v1 | DMT400001134 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
All Microarray Probes Designed to Gene DMG401000428
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18800_PI426222305 | JHI_St_60k_v1 | DMT400001138 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18847_PI426222305 | JHI_St_60k_v1 | DMT400001137 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18686_PI426222305 | JHI_St_60k_v1 | DMT400001140 | CGCCAAATATTGGCATAGTAAAGGTATTCTTGGTAGTCCTTTAATCAAATCTCATCTAAG |
| CUST_18731_PI426222305 | JHI_St_60k_v1 | DMT400001134 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18748_PI426222305 | JHI_St_60k_v1 | DMT400001136 | ATAGTTTTGCACTCAGTACAGTTTTTGCTATTGTAGGGTGGTGCTGCTGTCTGGTGCATG |
Microarray Signals from CUST_18731_PI426222305
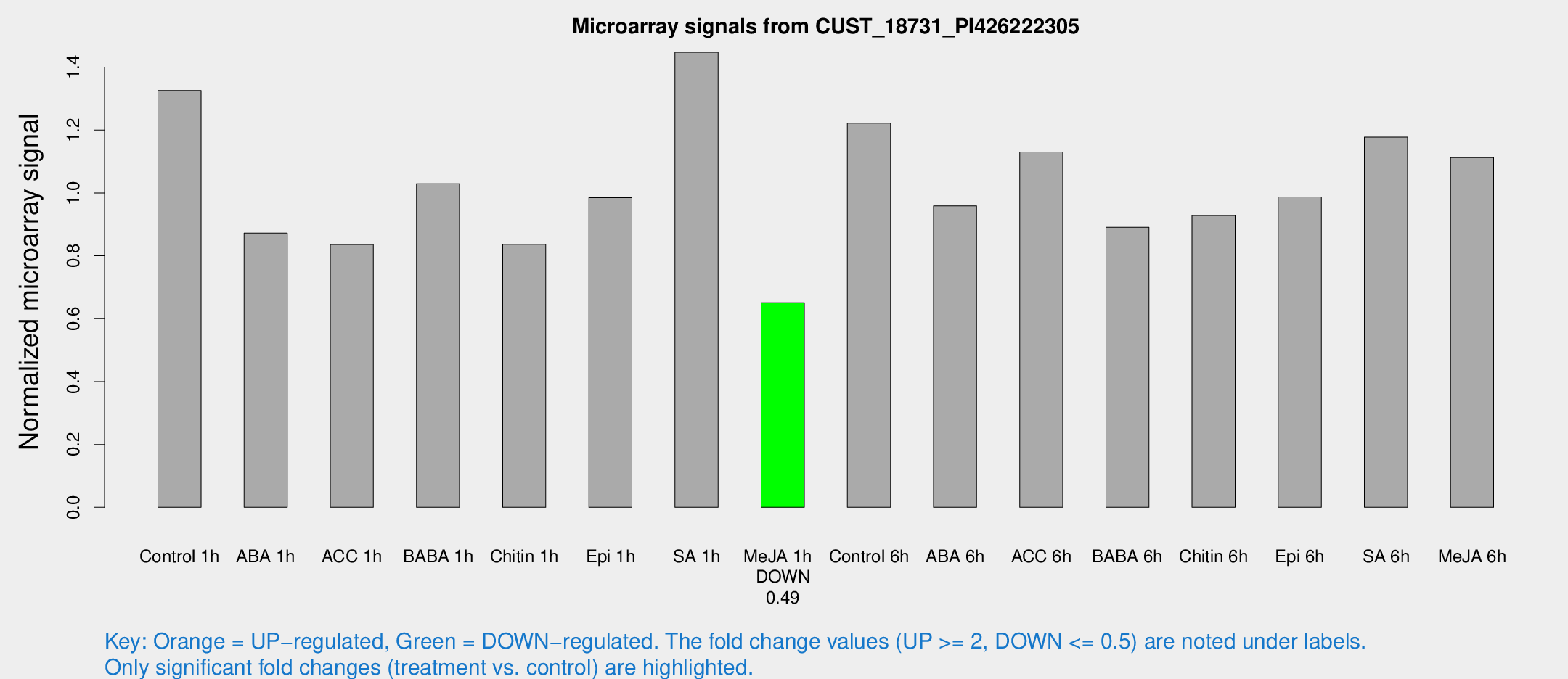
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2426.92 | 230 | 1.32578 | 0.0765658 |
| ABA 1h | 1401.2 | 81.059 | 0.872124 | 0.0503907 |
| ACC 1h | 1777.59 | 537.947 | 0.835892 | 0.286201 |
| BABA 1h | 1873.66 | 359.537 | 1.02953 | 0.11443 |
| Chitin 1h | 1370.04 | 90.4647 | 0.836346 | 0.0483277 |
| Epi 1h | 1562.85 | 153.369 | 0.98511 | 0.0897338 |
| SA 1h | 2754.92 | 379.366 | 1.44751 | 0.154146 |
| Me-JA 1h | 980.075 | 128.731 | 0.650632 | 0.0376343 |
| Control 6h | 2298.79 | 416.934 | 1.22216 | 0.150658 |
| ABA 6h | 1872.85 | 209.179 | 0.958995 | 0.0553979 |
| ACC 6h | 2354.4 | 136.167 | 1.12996 | 0.153097 |
| BABA 6h | 1824.78 | 164.658 | 0.890885 | 0.054765 |
| Chitin 6h | 1800.38 | 120.14 | 0.928244 | 0.0536278 |
| Epi 6h | 2027.37 | 127.127 | 0.987246 | 0.0570312 |
| SA 6h | 2314.15 | 636.677 | 1.17748 | 0.245254 |
| Me-JA 6h | 2057.15 | 317.78 | 1.11232 | 0.079469 |
Source Transcript PGSC0003DMT400001134 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G22850.1 | +3 | 4e-128 | 392 | 197/276 (71%) | Eukaryotic aspartyl protease family protein | chr5:7633717-7636298 REVERSE LENGTH=493 |
