Probe CUST_18748_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18748_PI426222305 | JHI_St_60k_v1 | DMT400001136 | ATAGTTTTGCACTCAGTACAGTTTTTGCTATTGTAGGGTGGTGCTGCTGTCTGGTGCATG |
All Microarray Probes Designed to Gene DMG401000428
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18800_PI426222305 | JHI_St_60k_v1 | DMT400001138 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18847_PI426222305 | JHI_St_60k_v1 | DMT400001137 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18686_PI426222305 | JHI_St_60k_v1 | DMT400001140 | CGCCAAATATTGGCATAGTAAAGGTATTCTTGGTAGTCCTTTAATCAAATCTCATCTAAG |
| CUST_18731_PI426222305 | JHI_St_60k_v1 | DMT400001134 | GTCCTAGTTCCACATATGGTATACTCATGTAGTGGTTAAATTTCTTGTGAAGTAGATTTC |
| CUST_18748_PI426222305 | JHI_St_60k_v1 | DMT400001136 | ATAGTTTTGCACTCAGTACAGTTTTTGCTATTGTAGGGTGGTGCTGCTGTCTGGTGCATG |
Microarray Signals from CUST_18748_PI426222305
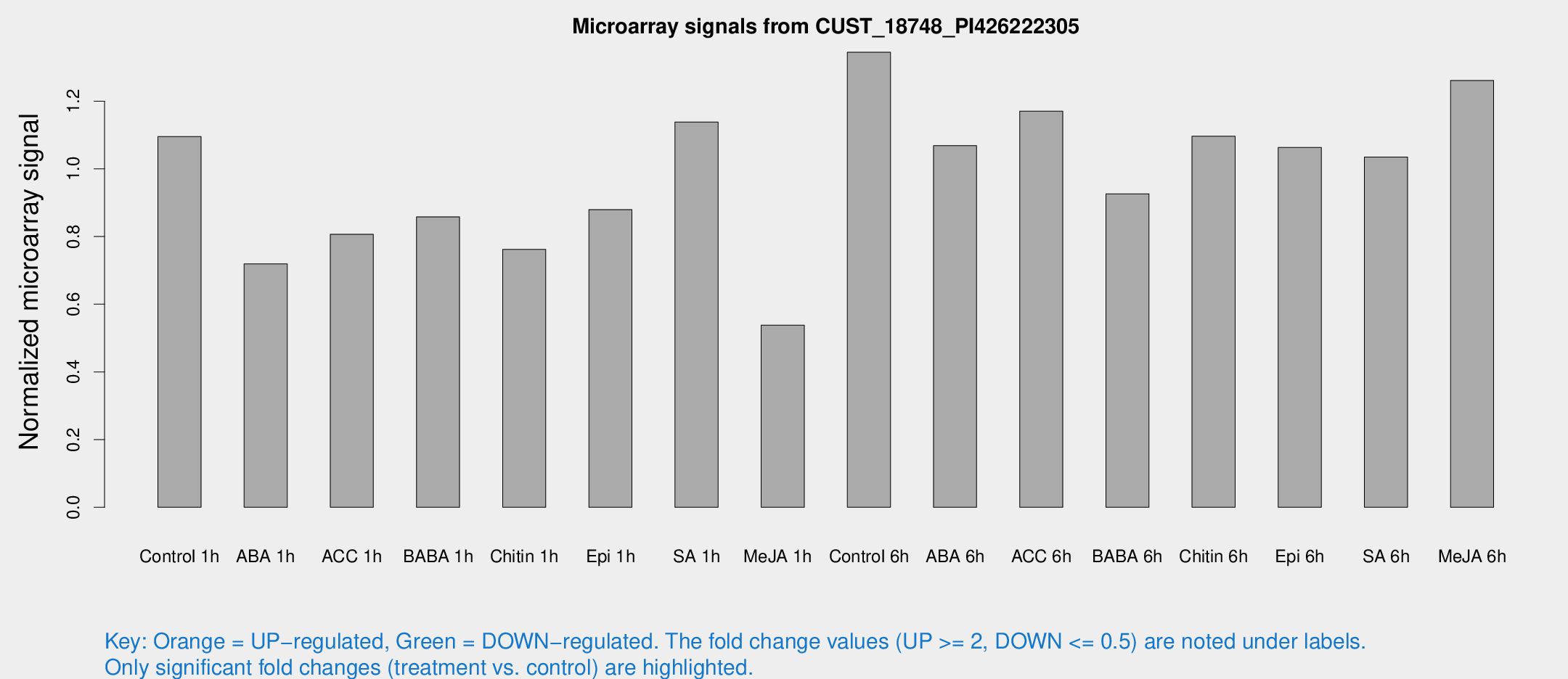
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 592.233 | 79.9676 | 1.09536 | 0.0889651 |
| ABA 1h | 338.839 | 24.4074 | 0.71895 | 0.0677192 |
| ACC 1h | 474.117 | 115.032 | 0.806727 | 0.178825 |
| BABA 1h | 453.561 | 78.8568 | 0.858188 | 0.076951 |
| Chitin 1h | 370.886 | 55.7194 | 0.761841 | 0.0582924 |
| Epi 1h | 405.846 | 30.2797 | 0.879543 | 0.0652081 |
| SA 1h | 645.555 | 122.054 | 1.13829 | 0.170739 |
| Me-JA 1h | 238.031 | 35.9852 | 0.538347 | 0.0373904 |
| Control 6h | 746.22 | 150.055 | 1.34447 | 0.190139 |
| ABA 6h | 626.068 | 115.982 | 1.06856 | 0.18627 |
| ACC 6h | 713.283 | 41.4958 | 1.17053 | 0.126069 |
| BABA 6h | 561.985 | 83.8762 | 0.926282 | 0.119437 |
| Chitin 6h | 623.3 | 55.2122 | 1.09623 | 0.0636983 |
| Epi 6h | 649.418 | 96.6954 | 1.06317 | 0.137245 |
| SA 6h | 553.216 | 73.2886 | 1.03476 | 0.060239 |
| Me-JA 6h | 673.76 | 74.0137 | 1.2611 | 0.0731557 |
Source Transcript PGSC0003DMT400001136 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G22850.1 | +2 | 9e-32 | 122 | 57/87 (66%) | Eukaryotic aspartyl protease family protein | chr5:7633717-7636298 REVERSE LENGTH=493 |
