Probe CUST_16819_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16819_PI426222305 | JHI_St_60k_v1 | DMT400069191 | GCGGTTGTATAAACTATGCGAAGTTGAATAAAATATTTTGGTCAAAAAATGTCCTTGCCG |
All Microarray Probes Designed to Gene DMG400026913
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16819_PI426222305 | JHI_St_60k_v1 | DMT400069191 | GCGGTTGTATAAACTATGCGAAGTTGAATAAAATATTTTGGTCAAAAAATGTCCTTGCCG |
| CUST_16564_PI426222305 | JHI_St_60k_v1 | DMT400069193 | GGTCCAACTTTAGTCTATCTATGTCTTTCGTTGGTTTTAACATCGAAATTCTACAGCATT |
| CUST_16622_PI426222305 | JHI_St_60k_v1 | DMT400069189 | GCAGCTGATTTCTCTACAGATGATATGATTCTTGAAAAGAAACTAGTTCCCCAGAGCTAG |
| CUST_16546_PI426222305 | JHI_St_60k_v1 | DMT400069190 | GGCTTAAGTTCTCTTTAAAGTTCAAATCATTTTTCGACTTCTACGTTGTCGTCCAAATTG |
Microarray Signals from CUST_16819_PI426222305
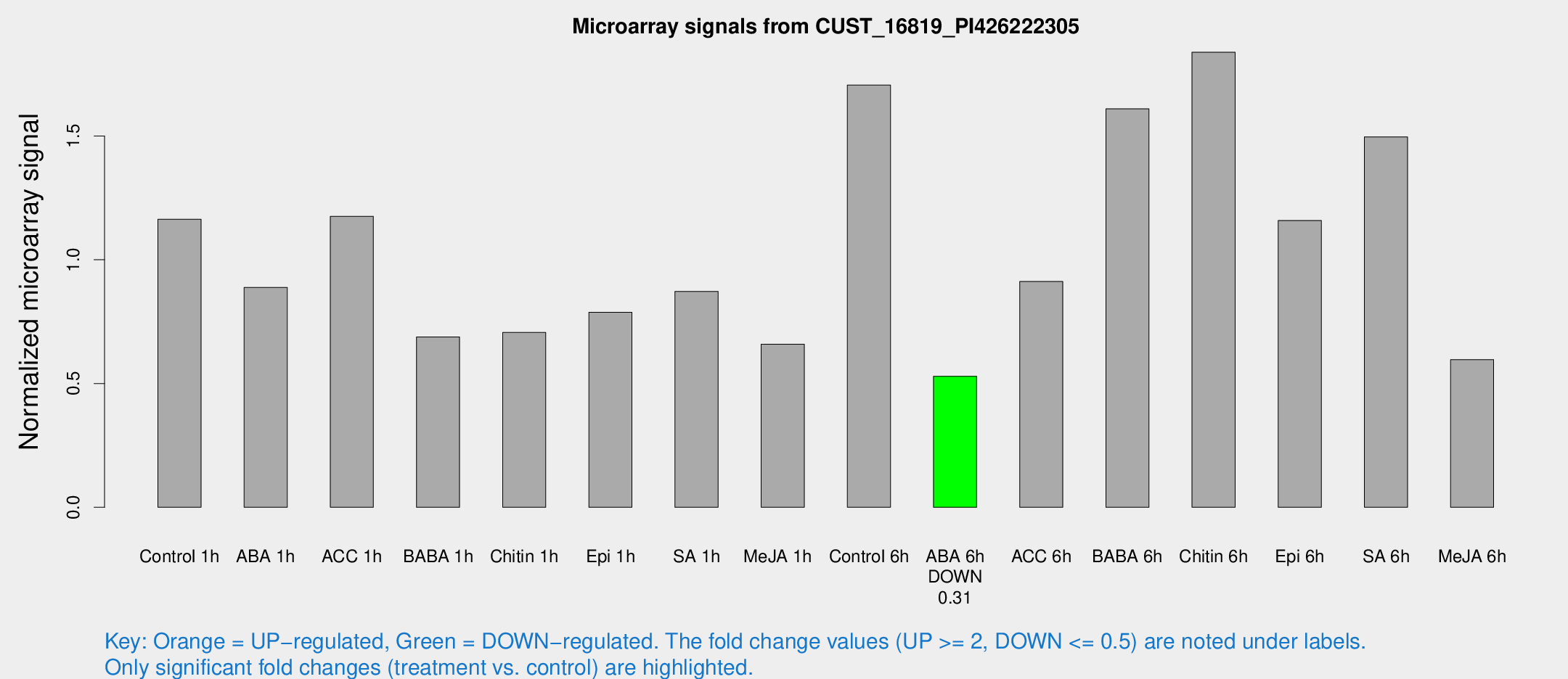
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 276.095 | 43.2272 | 1.16399 | 0.110758 |
| ABA 1h | 182.707 | 13.5066 | 0.88846 | 0.0533789 |
| ACC 1h | 280.495 | 19.6438 | 1.17554 | 0.176786 |
| BABA 1h | 162.337 | 34.7131 | 0.68829 | 0.0999527 |
| Chitin 1h | 153.002 | 31.2313 | 0.706333 | 0.133653 |
| Epi 1h | 161.755 | 26.6632 | 0.788102 | 0.10322 |
| SA 1h | 209.804 | 24.3643 | 0.871966 | 0.0520552 |
| Me-JA 1h | 125.784 | 13.0707 | 0.658603 | 0.0494072 |
| Control 6h | 418.094 | 96.3663 | 1.70561 | 0.297405 |
| ABA 6h | 130.583 | 8.27107 | 0.529494 | 0.0392925 |
| ACC 6h | 276.737 | 84.2407 | 0.912075 | 0.318922 |
| BABA 6h | 439.031 | 105.191 | 1.60978 | 0.359132 |
| Chitin 6h | 482.059 | 125.367 | 1.83848 | 0.409694 |
| Epi 6h | 303.538 | 20.1242 | 1.15891 | 0.146571 |
| SA 6h | 346.424 | 36.8132 | 1.49667 | 0.0877574 |
| Me-JA 6h | 163.259 | 62.3721 | 0.596602 | 0.227998 |
Source Transcript PGSC0003DMT400069191 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G09340.1 | +1 | 0.0 | 649 | 315/378 (83%) | chloroplast RNA binding | chr1:3015473-3018035 FORWARD LENGTH=378 |
