Probe CUST_16564_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16564_PI426222305 | JHI_St_60k_v1 | DMT400069193 | GGTCCAACTTTAGTCTATCTATGTCTTTCGTTGGTTTTAACATCGAAATTCTACAGCATT |
All Microarray Probes Designed to Gene DMG400026913
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16819_PI426222305 | JHI_St_60k_v1 | DMT400069191 | GCGGTTGTATAAACTATGCGAAGTTGAATAAAATATTTTGGTCAAAAAATGTCCTTGCCG |
| CUST_16564_PI426222305 | JHI_St_60k_v1 | DMT400069193 | GGTCCAACTTTAGTCTATCTATGTCTTTCGTTGGTTTTAACATCGAAATTCTACAGCATT |
| CUST_16622_PI426222305 | JHI_St_60k_v1 | DMT400069189 | GCAGCTGATTTCTCTACAGATGATATGATTCTTGAAAAGAAACTAGTTCCCCAGAGCTAG |
| CUST_16546_PI426222305 | JHI_St_60k_v1 | DMT400069190 | GGCTTAAGTTCTCTTTAAAGTTCAAATCATTTTTCGACTTCTACGTTGTCGTCCAAATTG |
Microarray Signals from CUST_16564_PI426222305
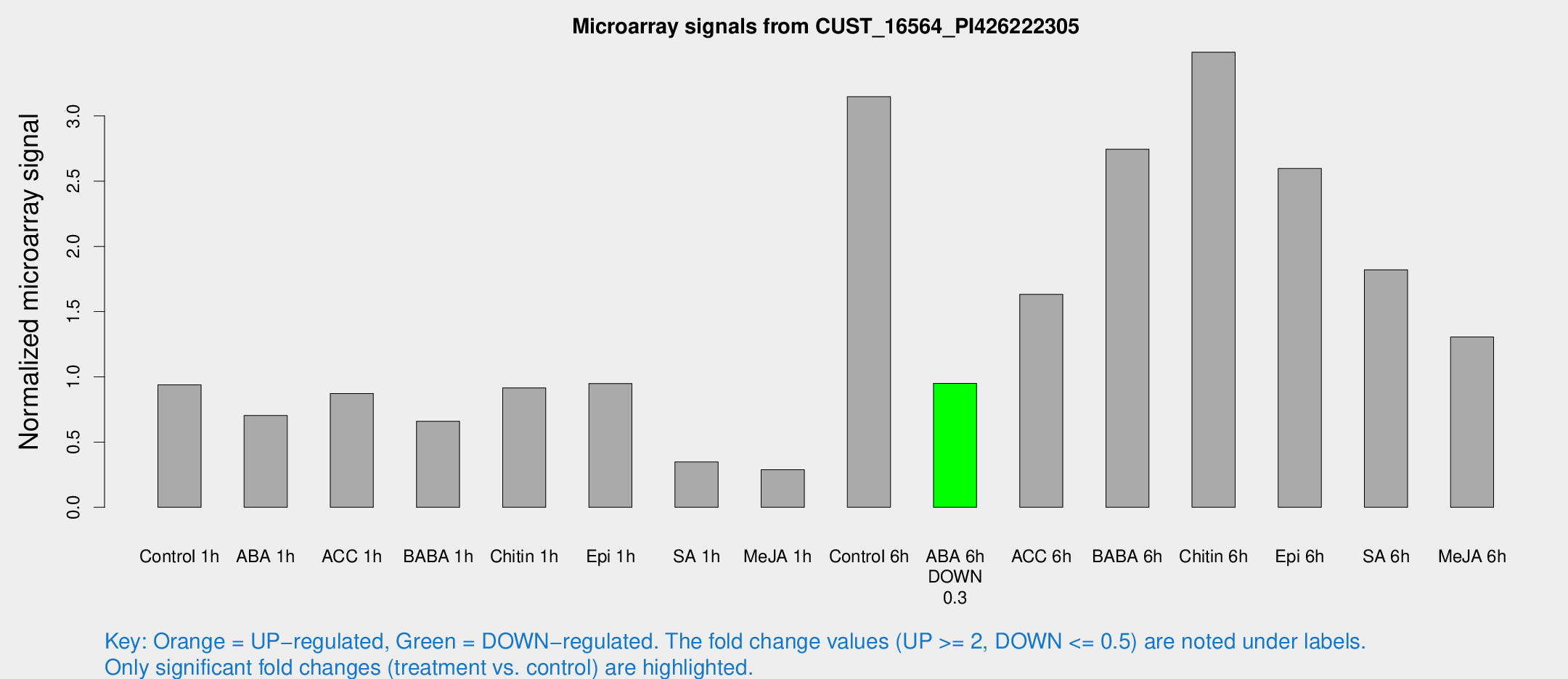
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 48.5994 | 4.05084 | 0.939492 | 0.0784646 |
| ABA 1h | 33.9179 | 7.70574 | 0.703773 | 0.129137 |
| ACC 1h | 47.6116 | 7.99969 | 0.872304 | 0.214045 |
| BABA 1h | 38.7415 | 14.2582 | 0.659717 | 0.243551 |
| Chitin 1h | 44.2723 | 9.51245 | 0.915585 | 0.184397 |
| Epi 1h | 42.4514 | 3.78517 | 0.948907 | 0.0855045 |
| SA 1h | 19.2731 | 4.26961 | 0.347806 | 0.0633901 |
| Me-JA 1h | 13.4017 | 4.37145 | 0.288755 | 0.0797938 |
| Control 6h | 181.428 | 60.7047 | 3.14846 | 0.9092 |
| ABA 6h | 52.2191 | 4.34161 | 0.950395 | 0.0802123 |
| ACC 6h | 111.302 | 39.6659 | 1.63261 | 0.478005 |
| BABA 6h | 164.679 | 32.1077 | 2.74546 | 0.498682 |
| Chitin 6h | 194.356 | 25.8179 | 3.48854 | 0.35141 |
| Epi 6h | 153.622 | 22.4601 | 2.59679 | 0.417472 |
| SA 6h | 103.754 | 36.7924 | 1.81983 | 0.520806 |
| Me-JA 6h | 92.5496 | 39.2751 | 1.30612 | 0.973617 |
Source Transcript PGSC0003DMT400069193 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G09340.1 | +2 | 4e-38 | 145 | 67/80 (84%) | chloroplast RNA binding | chr1:3015473-3018035 FORWARD LENGTH=378 |
