Probe CUST_16622_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16622_PI426222305 | JHI_St_60k_v1 | DMT400069189 | GCAGCTGATTTCTCTACAGATGATATGATTCTTGAAAAGAAACTAGTTCCCCAGAGCTAG |
All Microarray Probes Designed to Gene DMG400026913
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16819_PI426222305 | JHI_St_60k_v1 | DMT400069191 | GCGGTTGTATAAACTATGCGAAGTTGAATAAAATATTTTGGTCAAAAAATGTCCTTGCCG |
| CUST_16564_PI426222305 | JHI_St_60k_v1 | DMT400069193 | GGTCCAACTTTAGTCTATCTATGTCTTTCGTTGGTTTTAACATCGAAATTCTACAGCATT |
| CUST_16622_PI426222305 | JHI_St_60k_v1 | DMT400069189 | GCAGCTGATTTCTCTACAGATGATATGATTCTTGAAAAGAAACTAGTTCCCCAGAGCTAG |
| CUST_16546_PI426222305 | JHI_St_60k_v1 | DMT400069190 | GGCTTAAGTTCTCTTTAAAGTTCAAATCATTTTTCGACTTCTACGTTGTCGTCCAAATTG |
Microarray Signals from CUST_16622_PI426222305
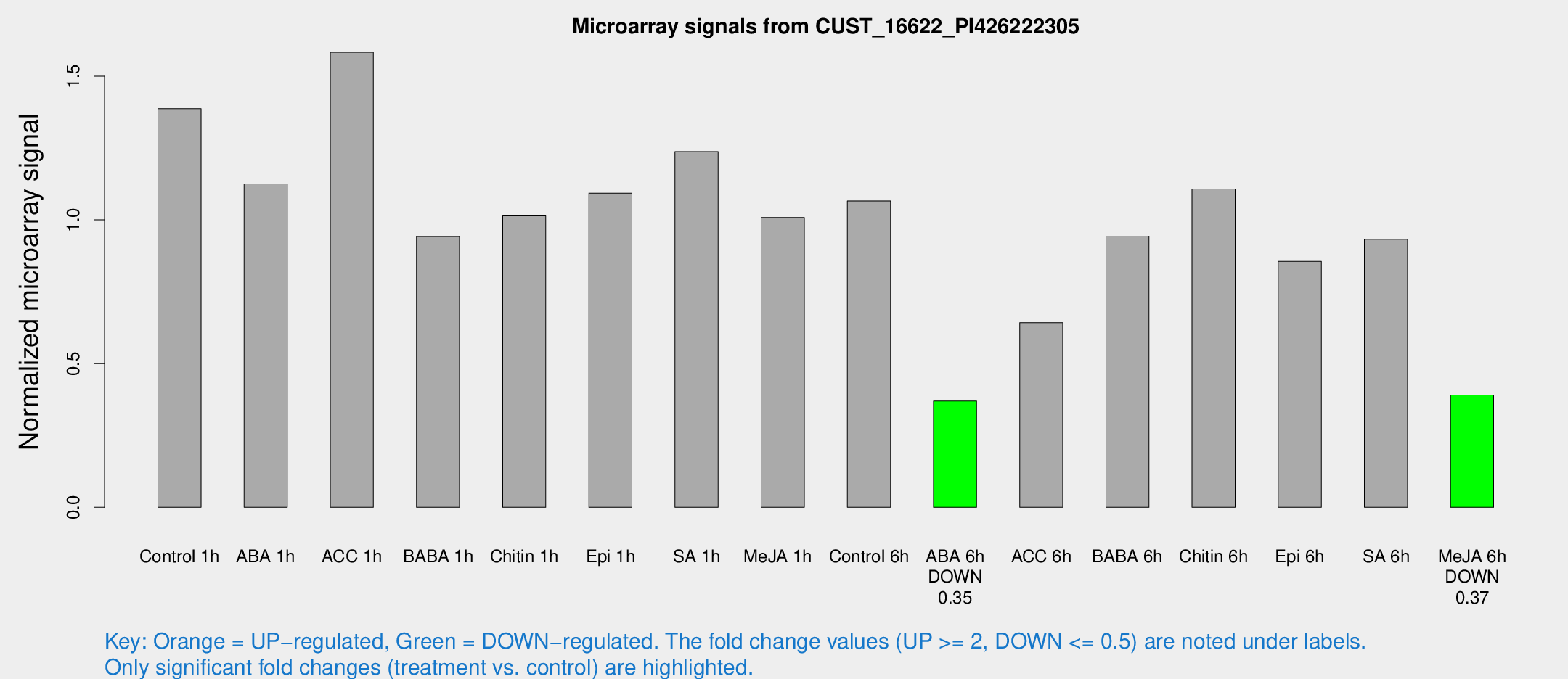
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 41375 | 5240.48 | 1.3869 | 0.100937 |
| ABA 1h | 29810.4 | 4102.39 | 1.12521 | 0.193724 |
| ACC 1h | 48174.1 | 4789.81 | 1.58325 | 0.268602 |
| BABA 1h | 27638.8 | 4941.04 | 0.942285 | 0.089919 |
| Chitin 1h | 27635.1 | 5214 | 1.01403 | 0.189341 |
| Epi 1h | 27885 | 2014.52 | 1.09321 | 0.0631166 |
| SA 1h | 37323.5 | 2157.99 | 1.23731 | 0.071436 |
| Me-JA 1h | 24349.9 | 2258.45 | 1.00825 | 0.0826285 |
| Control 6h | 33169.2 | 7279.8 | 1.06606 | 0.176634 |
| ABA 6h | 11563.3 | 669.697 | 0.369754 | 0.0229874 |
| ACC 6h | 22570.3 | 4662.13 | 0.642658 | 0.059429 |
| BABA 6h | 32011.4 | 5972.33 | 0.943484 | 0.182342 |
| Chitin 6h | 35112.1 | 4500.13 | 1.10772 | 0.103901 |
| Epi 6h | 28485.6 | 2272.4 | 0.855945 | 0.136759 |
| SA 6h | 27501.2 | 3411.19 | 0.932792 | 0.0538549 |
| Me-JA 6h | 12809.1 | 3930.05 | 0.390449 | 0.113538 |
Source Transcript PGSC0003DMT400069189 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G09340.1 | +1 | 0.0 | 633 | 315/409 (77%) | chloroplast RNA binding | chr1:3015473-3018035 FORWARD LENGTH=378 |
