Probe CUST_15312_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15312_PI426222305 | JHI_St_60k_v1 | DMT400057030 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
All Microarray Probes Designed to Gene DMG402022167
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14974_PI426222305 | JHI_St_60k_v1 | DMT400057033 | GCATATATGTAATTGTTGAAGCGTTTCACGTCGGATGAAATAGGGATATGTGGTTTTCTT |
| CUST_15312_PI426222305 | JHI_St_60k_v1 | DMT400057030 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
| CUST_15134_PI426222305 | JHI_St_60k_v1 | DMT400057028 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
Microarray Signals from CUST_15312_PI426222305
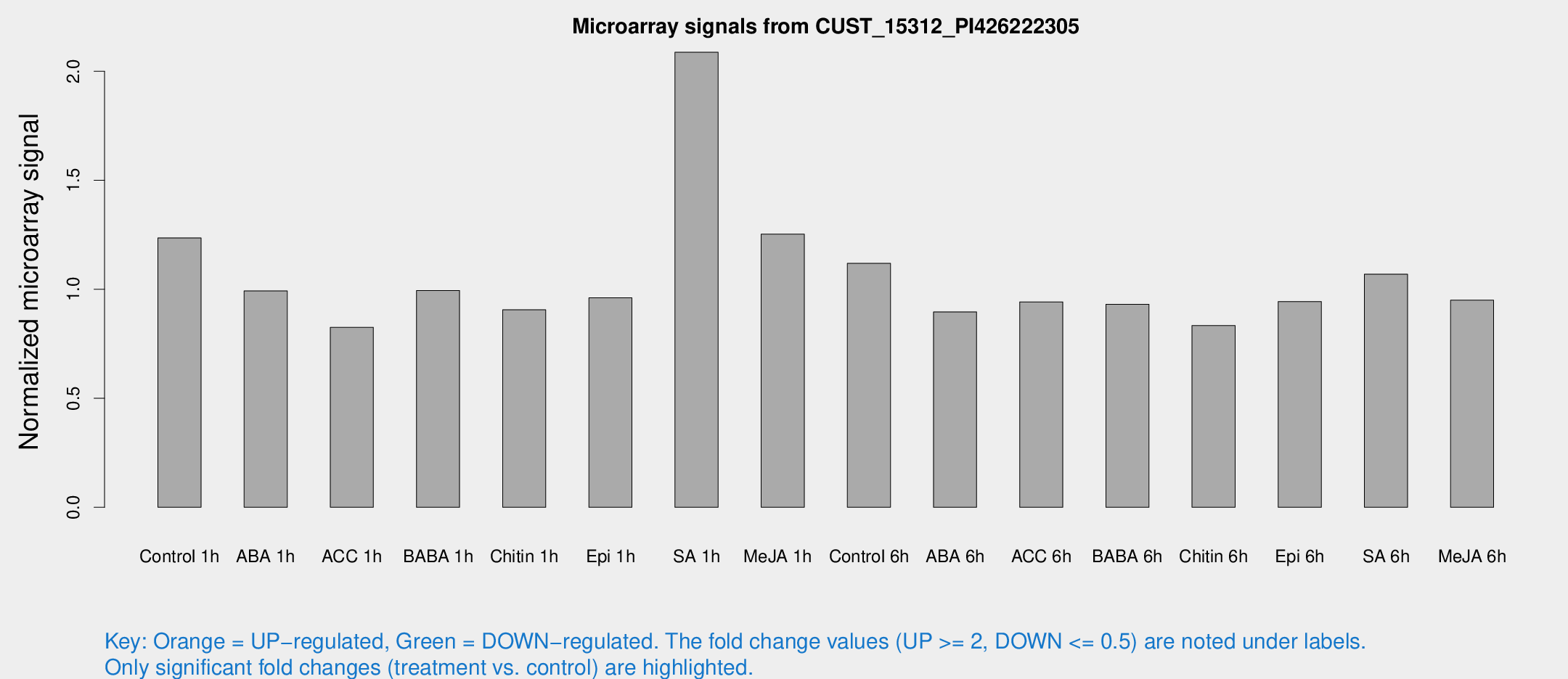
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3688.66 | 369.591 | 1.23553 | 0.0713448 |
| ABA 1h | 2620.11 | 253.319 | 0.992325 | 0.0573085 |
| ACC 1h | 2585.12 | 437.017 | 0.825167 | 0.0940038 |
| BABA 1h | 3024.51 | 689.145 | 0.994264 | 0.16567 |
| Chitin 1h | 2420.48 | 177.248 | 0.905925 | 0.0523219 |
| Epi 1h | 2473.64 | 197.515 | 0.960608 | 0.0757952 |
| SA 1h | 6395.06 | 584.592 | 2.08723 | 0.120512 |
| Me-JA 1h | 3072.25 | 396.728 | 1.25272 | 0.0723429 |
| Control 6h | 3463.86 | 707.958 | 1.11871 | 0.163009 |
| ABA 6h | 2878.67 | 409.756 | 0.896152 | 0.0889989 |
| ACC 6h | 3222.77 | 294.123 | 0.942119 | 0.0763541 |
| BABA 6h | 3099.73 | 237.463 | 0.931396 | 0.0591062 |
| Chitin 6h | 2646.19 | 258.852 | 0.833352 | 0.0583985 |
| Epi 6h | 3161.29 | 216.916 | 0.943801 | 0.0545068 |
| SA 6h | 3579.38 | 1090.72 | 1.06918 | 0.318593 |
| Me-JA 6h | 2835.36 | 333.845 | 0.949799 | 0.0548517 |
Source Transcript PGSC0003DMT400057030 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55850.1 | +1 | 0.0 | 827 | 404/747 (54%) | cellulose synthase like E1 | chr1:20876752-20879414 FORWARD LENGTH=729 |
