Probe CUST_14974_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14974_PI426222305 | JHI_St_60k_v1 | DMT400057033 | GCATATATGTAATTGTTGAAGCGTTTCACGTCGGATGAAATAGGGATATGTGGTTTTCTT |
All Microarray Probes Designed to Gene DMG402022167
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14974_PI426222305 | JHI_St_60k_v1 | DMT400057033 | GCATATATGTAATTGTTGAAGCGTTTCACGTCGGATGAAATAGGGATATGTGGTTTTCTT |
| CUST_15312_PI426222305 | JHI_St_60k_v1 | DMT400057030 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
| CUST_15134_PI426222305 | JHI_St_60k_v1 | DMT400057028 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
Microarray Signals from CUST_14974_PI426222305
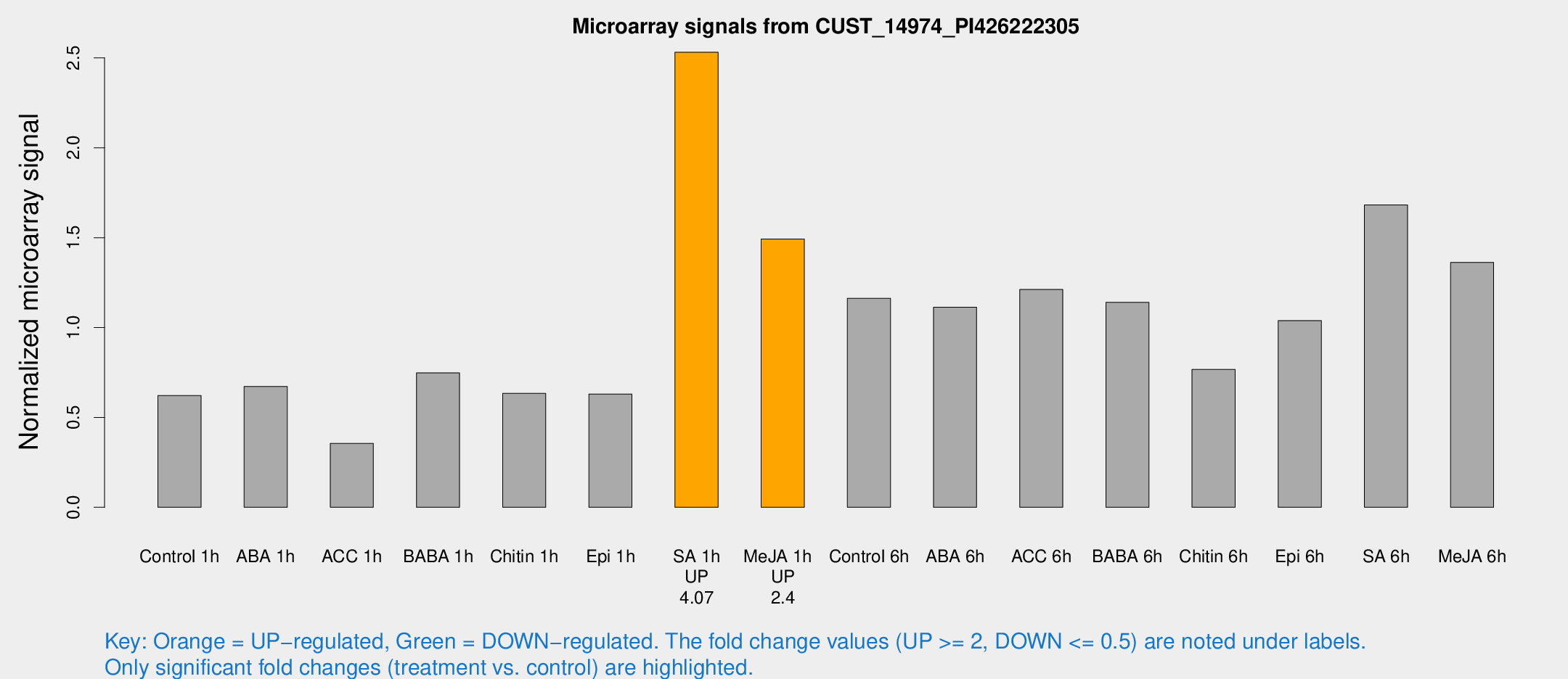
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 292.051 | 17.2776 | 0.621692 | 0.0366815 |
| ABA 1h | 285.472 | 43.0788 | 0.672015 | 0.0852629 |
| ACC 1h | 194.976 | 62.9956 | 0.355272 | 0.119926 |
| BABA 1h | 355.157 | 72.7233 | 0.747663 | 0.0984423 |
| Chitin 1h | 271.532 | 35.537 | 0.633891 | 0.0703503 |
| Epi 1h | 259.914 | 33.1372 | 0.630093 | 0.0729246 |
| SA 1h | 1225.34 | 93.0738 | 2.53158 | 0.146351 |
| Me-JA 1h | 581.779 | 82.4326 | 1.49265 | 0.0867199 |
| Control 6h | 564.984 | 103.907 | 1.16256 | 0.14681 |
| ABA 6h | 576.319 | 111.136 | 1.11311 | 0.16432 |
| ACC 6h | 656.111 | 55.5338 | 1.212 | 0.145534 |
| BABA 6h | 603.57 | 59.4403 | 1.14024 | 0.0682423 |
| Chitin 6h | 384.548 | 28.5989 | 0.766959 | 0.05675 |
| Epi 6h | 559.674 | 76.3899 | 1.03863 | 0.0733998 |
| SA 6h | 848.024 | 218.26 | 1.68183 | 0.330661 |
| Me-JA 6h | 644.518 | 77.7293 | 1.36244 | 0.0790656 |
Source Transcript PGSC0003DMT400057033 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55850.1 | +1 | 1e-144 | 436 | 207/388 (53%) | cellulose synthase like E1 | chr1:20876752-20879414 FORWARD LENGTH=729 |
