Probe CUST_15134_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_15134_PI426222305 | JHI_St_60k_v1 | DMT400057028 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
All Microarray Probes Designed to Gene DMG402022167
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14974_PI426222305 | JHI_St_60k_v1 | DMT400057033 | GCATATATGTAATTGTTGAAGCGTTTCACGTCGGATGAAATAGGGATATGTGGTTTTCTT |
| CUST_15312_PI426222305 | JHI_St_60k_v1 | DMT400057030 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
| CUST_15134_PI426222305 | JHI_St_60k_v1 | DMT400057028 | TTGCCTTTGTATAATGCTCTCTTCTTTAGGCAAGATAAGGGGAAAATACCTAGTTCCACT |
Microarray Signals from CUST_15134_PI426222305
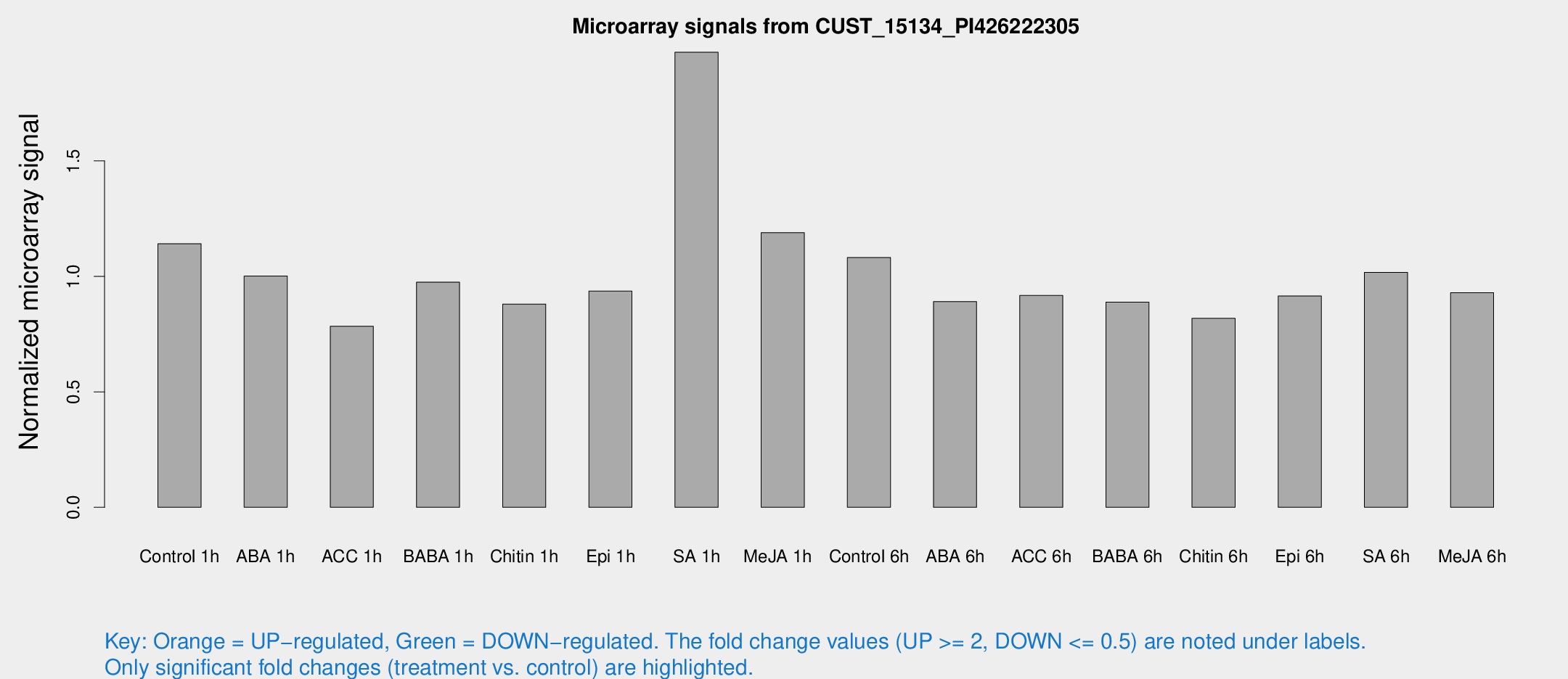
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3600.54 | 381.326 | 1.1414 | 0.0659055 |
| ABA 1h | 2793.91 | 274.488 | 1.00151 | 0.0578318 |
| ACC 1h | 2638.71 | 550.328 | 0.784375 | 0.126127 |
| BABA 1h | 3090.07 | 638.873 | 0.975059 | 0.130937 |
| Chitin 1h | 2515.07 | 350.198 | 0.880006 | 0.0743521 |
| Epi 1h | 2543.28 | 181.304 | 0.936417 | 0.0559844 |
| SA 1h | 6434 | 825.708 | 1.97072 | 0.159472 |
| Me-JA 1h | 3095.61 | 465.239 | 1.18939 | 0.0804871 |
| Control 6h | 3555.37 | 752.719 | 1.08147 | 0.166063 |
| ABA 6h | 3023.45 | 433.31 | 0.891015 | 0.0904732 |
| ACC 6h | 3313.68 | 304.768 | 0.917291 | 0.0742184 |
| BABA 6h | 3126.1 | 255 | 0.888745 | 0.0545985 |
| Chitin 6h | 2743.9 | 269.507 | 0.818297 | 0.0589795 |
| Epi 6h | 3239.36 | 246.478 | 0.915173 | 0.0657412 |
| SA 6h | 3649.88 | 1154.95 | 1.01706 | 0.332985 |
| Me-JA 6h | 2941.54 | 399.319 | 0.929403 | 0.0536681 |
Source Transcript PGSC0003DMT400057028 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55850.1 | +1 | 5e-137 | 436 | 207/388 (53%) | cellulose synthase like E1 | chr1:20876752-20879414 FORWARD LENGTH=729 |
