Probe CUST_12916_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12916_PI426222305 | JHI_St_60k_v1 | DMT400062881 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
All Microarray Probes Designed to Gene DMG400024475
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12775_PI426222305 | JHI_St_60k_v1 | DMT400062879 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12916_PI426222305 | JHI_St_60k_v1 | DMT400062881 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12943_PI426222305 | JHI_St_60k_v1 | DMT400062878 | TGTATCTGGTCCAAAAACCGGTGTTGTTTTTAACATTGTTGGACAAGATGATGCTCGTAA |
| CUST_12896_PI426222305 | JHI_St_60k_v1 | DMT400062880 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
Microarray Signals from CUST_12916_PI426222305
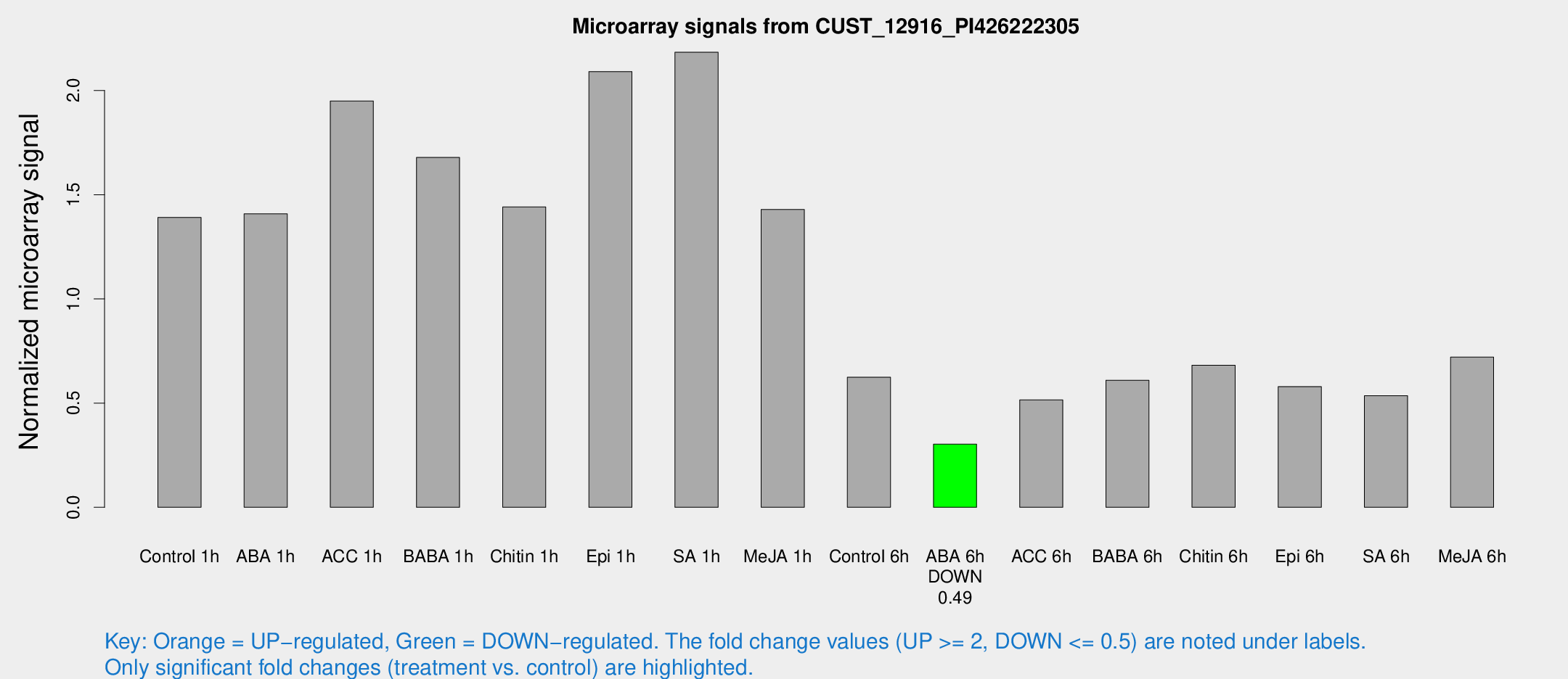
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 943.23 | 115.918 | 1.39098 | 0.111455 |
| ABA 1h | 837.739 | 75.42 | 1.40824 | 0.187024 |
| ACC 1h | 1425.53 | 326.104 | 1.9494 | 0.379977 |
| BABA 1h | 1092.48 | 124.098 | 1.67873 | 0.139674 |
| Chitin 1h | 867.637 | 55.318 | 1.44103 | 0.106697 |
| Epi 1h | 1210.85 | 70.2079 | 2.09103 | 0.120862 |
| SA 1h | 1521.29 | 190.966 | 2.18375 | 0.217642 |
| Me-JA 1h | 778.301 | 45.0829 | 1.42885 | 0.152636 |
| Control 6h | 427.616 | 63.7659 | 0.624085 | 0.0484524 |
| ABA 6h | 215.499 | 12.9607 | 0.303003 | 0.0181781 |
| ACC 6h | 400.638 | 51.882 | 0.51504 | 0.0419424 |
| BABA 6h | 462.189 | 55.3966 | 0.609513 | 0.0575697 |
| Chitin 6h | 484.662 | 28.2784 | 0.681453 | 0.0396922 |
| Epi 6h | 441.228 | 50.8009 | 0.579347 | 0.0623291 |
| SA 6h | 365.402 | 65.0239 | 0.535551 | 0.0463314 |
| Me-JA 6h | 491.369 | 74.9369 | 0.720616 | 0.0646644 |
Source Transcript PGSC0003DMT400062881 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62570.2 | +3 | 4e-152 | 454 | 256/527 (49%) | Calmodulin binding protein-like | chr5:25114934-25116967 FORWARD LENGTH=494 |
