Probe CUST_12775_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12775_PI426222305 | JHI_St_60k_v1 | DMT400062879 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
All Microarray Probes Designed to Gene DMG400024475
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12775_PI426222305 | JHI_St_60k_v1 | DMT400062879 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12916_PI426222305 | JHI_St_60k_v1 | DMT400062881 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12943_PI426222305 | JHI_St_60k_v1 | DMT400062878 | TGTATCTGGTCCAAAAACCGGTGTTGTTTTTAACATTGTTGGACAAGATGATGCTCGTAA |
| CUST_12896_PI426222305 | JHI_St_60k_v1 | DMT400062880 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
Microarray Signals from CUST_12775_PI426222305
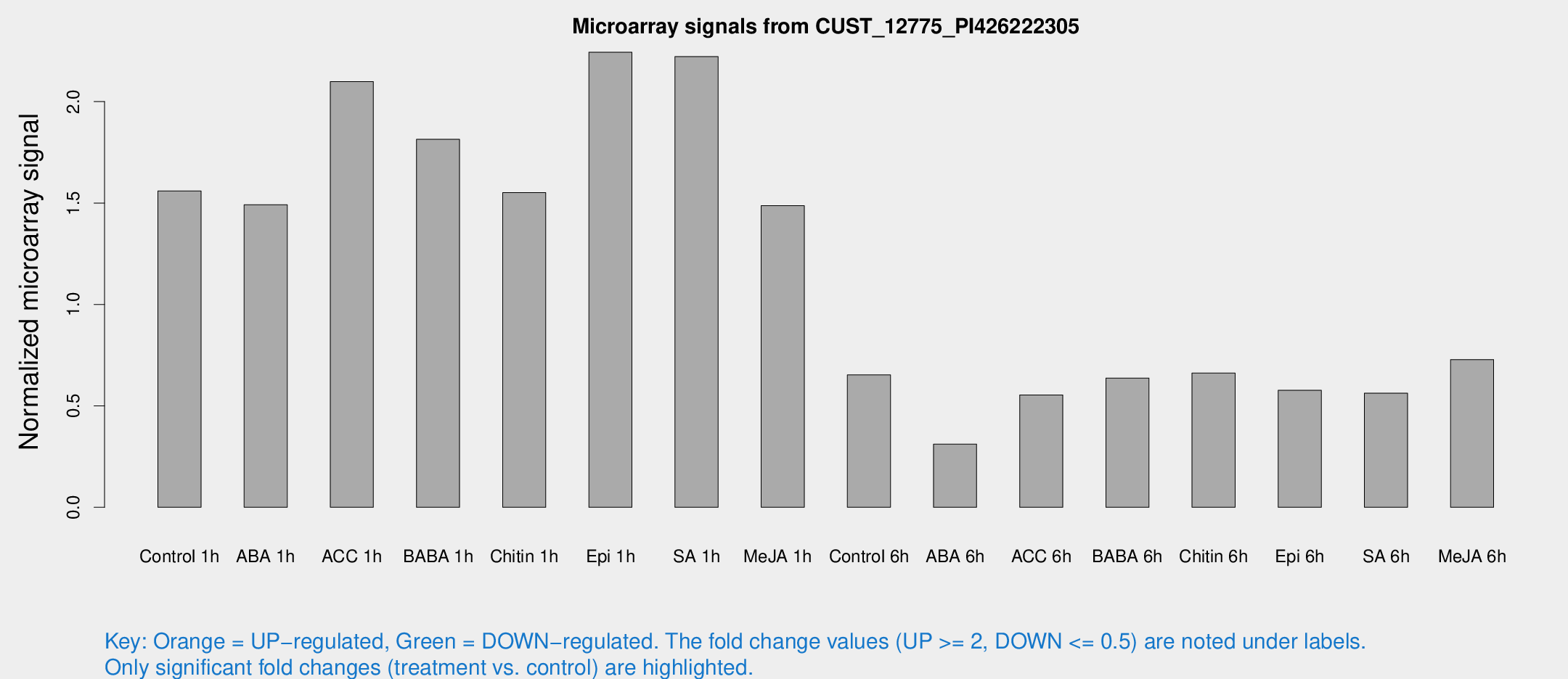
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1051.82 | 93.8615 | 1.55967 | 0.0901516 |
| ABA 1h | 894.98 | 102.634 | 1.49196 | 0.231733 |
| ACC 1h | 1547.35 | 366.413 | 2.09886 | 0.435046 |
| BABA 1h | 1186.05 | 141.36 | 1.81436 | 0.120801 |
| Chitin 1h | 935.01 | 54.1841 | 1.55144 | 0.12621 |
| Epi 1h | 1301.17 | 75.3256 | 2.24365 | 0.129635 |
| SA 1h | 1552.72 | 194.878 | 2.2219 | 0.213235 |
| Me-JA 1h | 813.419 | 47.1484 | 1.48707 | 0.193585 |
| Control 6h | 451.239 | 73.5338 | 0.652825 | 0.0567001 |
| ABA 6h | 226.08 | 33.9451 | 0.311373 | 0.0288281 |
| ACC 6h | 432.028 | 56.8379 | 0.554029 | 0.0323138 |
| BABA 6h | 488.143 | 72.6479 | 0.637244 | 0.0816434 |
| Chitin 6h | 475.723 | 48.5735 | 0.661693 | 0.0458485 |
| Epi 6h | 437.268 | 32.5623 | 0.576816 | 0.0644173 |
| SA 6h | 385.876 | 71.9922 | 0.562508 | 0.0549946 |
| Me-JA 6h | 492.794 | 60.3095 | 0.728434 | 0.0423119 |
Source Transcript PGSC0003DMT400062879 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62570.2 | +3 | 2e-103 | 335 | 168/269 (62%) | Calmodulin binding protein-like | chr5:25114934-25116967 FORWARD LENGTH=494 |
