Probe CUST_12896_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12896_PI426222305 | JHI_St_60k_v1 | DMT400062880 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
All Microarray Probes Designed to Gene DMG400024475
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_12775_PI426222305 | JHI_St_60k_v1 | DMT400062879 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12916_PI426222305 | JHI_St_60k_v1 | DMT400062881 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
| CUST_12943_PI426222305 | JHI_St_60k_v1 | DMT400062878 | TGTATCTGGTCCAAAAACCGGTGTTGTTTTTAACATTGTTGGACAAGATGATGCTCGTAA |
| CUST_12896_PI426222305 | JHI_St_60k_v1 | DMT400062880 | CAGCAACCTTTGCTGCATTACTTATTTTGTGCATGTGCATATTTCCAATTTTATCTTGTG |
Microarray Signals from CUST_12896_PI426222305
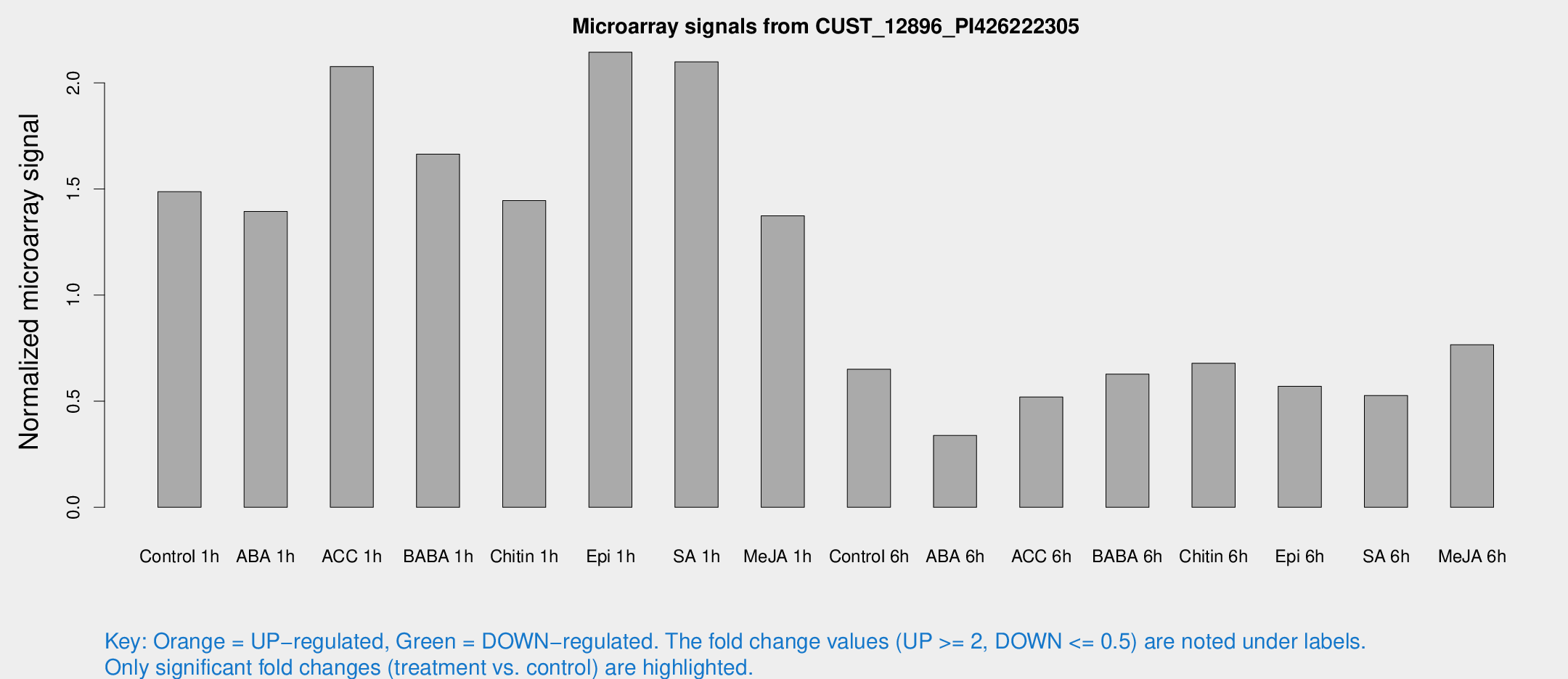
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1012.58 | 85.851 | 1.48728 | 0.0859891 |
| ABA 1h | 841.375 | 80.5064 | 1.39403 | 0.180174 |
| ACC 1h | 1516.94 | 309.901 | 2.07725 | 0.333418 |
| BABA 1h | 1098.3 | 126.593 | 1.66434 | 0.104028 |
| Chitin 1h | 878.667 | 50.8591 | 1.44519 | 0.0928836 |
| Epi 1h | 1259.38 | 80.4937 | 2.14444 | 0.123921 |
| SA 1h | 1470.56 | 136.552 | 2.09943 | 0.132069 |
| Me-JA 1h | 757.984 | 43.9035 | 1.37319 | 0.132585 |
| Control 6h | 455.094 | 76.5511 | 0.650333 | 0.0597724 |
| ABA 6h | 245.52 | 24.0608 | 0.338667 | 0.0214859 |
| ACC 6h | 412.313 | 62.7269 | 0.519938 | 0.0443126 |
| BABA 6h | 482.464 | 58.7466 | 0.627603 | 0.0603655 |
| Chitin 6h | 494.036 | 53.4339 | 0.679217 | 0.050528 |
| Epi 6h | 436.388 | 30.9957 | 0.570063 | 0.0561496 |
| SA 6h | 364.014 | 64.069 | 0.527084 | 0.0449789 |
| Me-JA 6h | 525.79 | 69.4033 | 0.76638 | 0.0546349 |
Source Transcript PGSC0003DMT400062880 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62570.2 | +3 | 3e-150 | 454 | 256/527 (49%) | Calmodulin binding protein-like | chr5:25114934-25116967 FORWARD LENGTH=494 |
