Probe CUST_1153_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1153_PI426222305 | JHI_St_60k_v1 | DMT400032815 | AACTGACTGGTGTAATGTTCTAAAATTTTGTTTGCTGAGGCCAATGGAGTTGAATATAAC |
All Microarray Probes Designed to Gene DMG400012603
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_782_PI426222305 | JHI_St_60k_v1 | DMT400032814 | AAGACCCCCGGAGTTGATGTGAAGCCTTCAGGTAATTATTTTGAGATTGATTTCTTCTGA |
| CUST_1153_PI426222305 | JHI_St_60k_v1 | DMT400032815 | AACTGACTGGTGTAATGTTCTAAAATTTTGTTTGCTGAGGCCAATGGAGTTGAATATAAC |
| CUST_1106_PI426222305 | JHI_St_60k_v1 | DMT400032816 | GCCTTGATGTGGCTATAAAGCTAGGTATTGCTGGATGTGATGTTTTAAAATTTTCAATTG |
Microarray Signals from CUST_1153_PI426222305
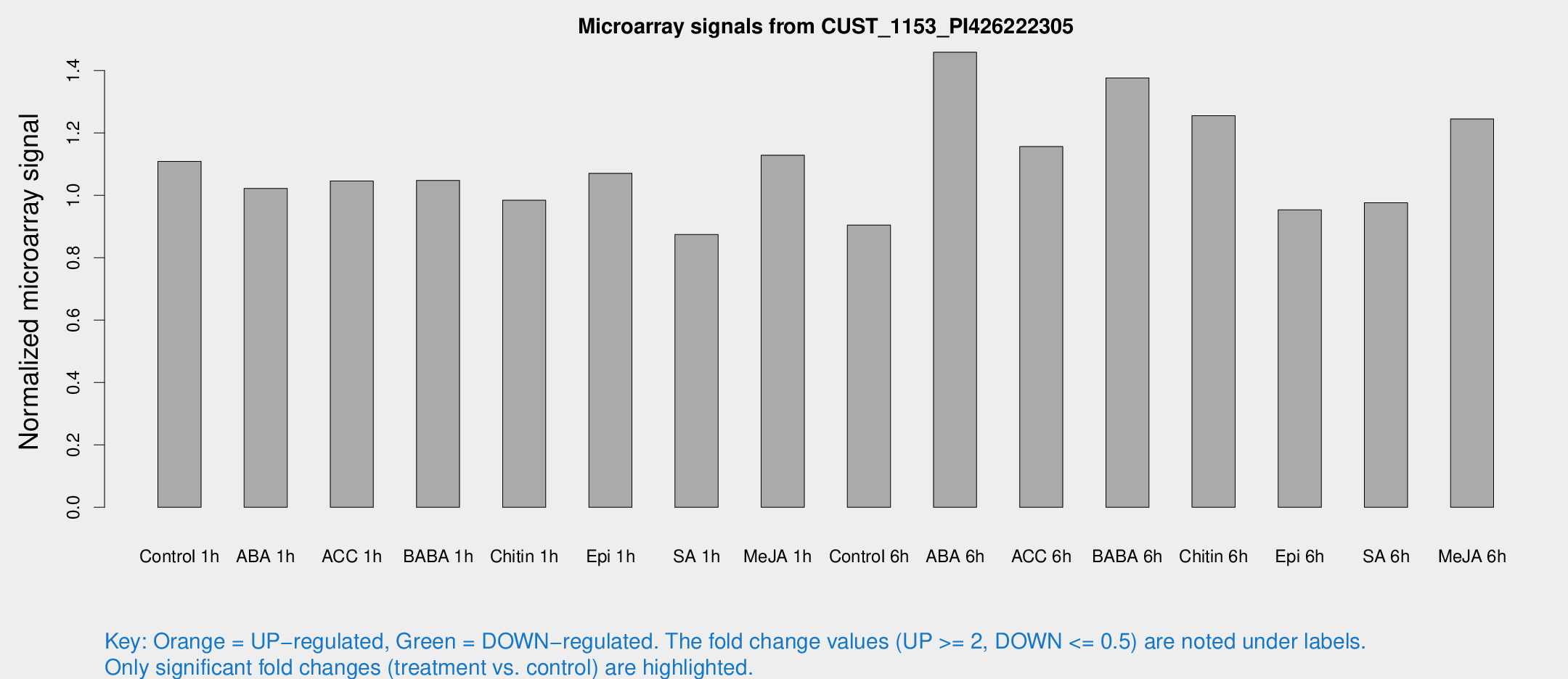
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.5094 | 3.4633 | 1.10894 | 0.529964 |
| ABA 1h | 6.04857 | 3.34724 | 1.02198 | 0.566886 |
| ACC 1h | 7.36474 | 4.41223 | 1.04587 | 0.607276 |
| BABA 1h | 6.75222 | 3.63505 | 1.0478 | 0.568547 |
| Chitin 1h | 5.90745 | 3.43166 | 0.984292 | 0.570325 |
| Epi 1h | 6.17617 | 3.37398 | 1.07063 | 0.586148 |
| SA 1h | 5.98238 | 3.47037 | 0.874447 | 0.507107 |
| Me-JA 1h | 6.14641 | 3.56643 | 1.12858 | 0.653463 |
| Control 6h | 6.03845 | 3.49778 | 0.904818 | 0.52401 |
| ABA 6h | 10.7999 | 3.66294 | 1.45864 | 0.559134 |
| ACC 6h | 9.0753 | 4.41616 | 1.15626 | 0.573521 |
| BABA 6h | 11.3802 | 3.96747 | 1.37585 | 0.577154 |
| Chitin 6h | 9.58811 | 3.91195 | 1.25497 | 0.600914 |
| Epi 6h | 7.21281 | 4.20997 | 0.953464 | 0.552109 |
| SA 6h | 6.43516 | 3.73149 | 0.976373 | 0.565632 |
| Me-JA 6h | 9.15251 | 3.56627 | 1.24478 | 0.591244 |
Source Transcript PGSC0003DMT400032815 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g093360.2 | +1 | 0.0 | 607 | 328/351 (93%) | genomic_reference:SL2.50ch02 gene_region:48831308-48835273 transcript_region:SL2.50ch02:48831308..48835273+ go_terms:GO:0004711 functional_description:Ribosomal protein S6 kinase alpha-3 (AHRD V1 *-** KS6A3_HUMAN); contains Interpro domain(s) IPR002290 Serine/threonine protein kinase |
| TAIR PP10 | AT5G47750.1 | +1 | 2e-67 | 233 | 133/199 (67%) | D6 protein kinase like 2 | chr5:19339947-19341864 REVERSE LENGTH=586 |
