Probe CUST_782_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_782_PI426222305 | JHI_St_60k_v1 | DMT400032814 | AAGACCCCCGGAGTTGATGTGAAGCCTTCAGGTAATTATTTTGAGATTGATTTCTTCTGA |
All Microarray Probes Designed to Gene DMG400012603
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_782_PI426222305 | JHI_St_60k_v1 | DMT400032814 | AAGACCCCCGGAGTTGATGTGAAGCCTTCAGGTAATTATTTTGAGATTGATTTCTTCTGA |
| CUST_1153_PI426222305 | JHI_St_60k_v1 | DMT400032815 | AACTGACTGGTGTAATGTTCTAAAATTTTGTTTGCTGAGGCCAATGGAGTTGAATATAAC |
| CUST_1106_PI426222305 | JHI_St_60k_v1 | DMT400032816 | GCCTTGATGTGGCTATAAAGCTAGGTATTGCTGGATGTGATGTTTTAAAATTTTCAATTG |
Microarray Signals from CUST_782_PI426222305
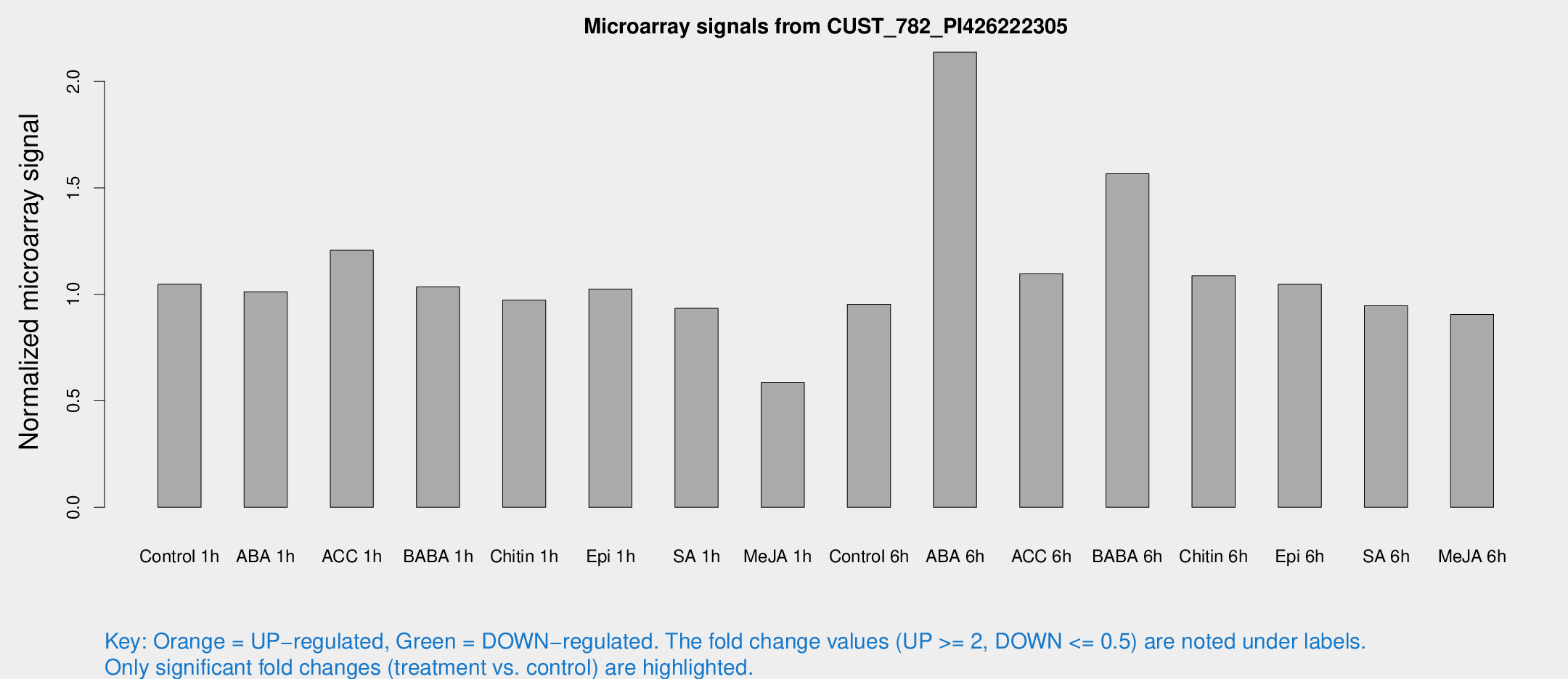
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 812.312 | 91.1961 | 1.04733 | 0.0625511 |
| ABA 1h | 699.166 | 97.127 | 1.01204 | 0.0676982 |
| ACC 1h | 980.645 | 165.901 | 1.20667 | 0.127396 |
| BABA 1h | 809.732 | 191.888 | 1.03475 | 0.166151 |
| Chitin 1h | 675.271 | 55.8116 | 0.972734 | 0.056353 |
| Epi 1h | 693.103 | 92.0055 | 1.02438 | 0.135054 |
| SA 1h | 740.409 | 58.9145 | 0.934055 | 0.0540818 |
| Me-JA 1h | 370.806 | 41.7981 | 0.585522 | 0.0370653 |
| Control 6h | 811.622 | 223.545 | 0.953363 | 0.250797 |
| ABA 6h | 1755.44 | 153.894 | 2.13675 | 0.123439 |
| ACC 6h | 998.778 | 192.685 | 1.09637 | 0.0963832 |
| BABA 6h | 1358.14 | 145.658 | 1.56584 | 0.124591 |
| Chitin 6h | 904.493 | 128.692 | 1.08781 | 0.150636 |
| Epi 6h | 920.247 | 116.515 | 1.04719 | 0.100167 |
| SA 6h | 739.322 | 118.858 | 0.946743 | 0.0558142 |
| Me-JA 6h | 732.91 | 167.808 | 0.905583 | 0.182654 |
Source Transcript PGSC0003DMT400032814 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g093360.2 | +1 | 0.0 | 1016 | 562/632 (89%) | genomic_reference:SL2.50ch02 gene_region:48831308-48835273 transcript_region:SL2.50ch02:48831308..48835273+ go_terms:GO:0004711 functional_description:Ribosomal protein S6 kinase alpha-3 (AHRD V1 *-** KS6A3_HUMAN); contains Interpro domain(s) IPR002290 Serine/threonine protein kinase |
| TAIR PP10 | AT3G27580.1 | +1 | 0.0 | 528 | 337/599 (56%) | Protein kinase superfamily protein | chr3:10217671-10219484 REVERSE LENGTH=578 |
