Probe CUST_33815_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33815_PI426222305 | JHI_St_60k_v1 | DMT400070920 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
All Microarray Probes Designed to Gene DMG400027577
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33811_PI426222305 | JHI_St_60k_v1 | DMT400070918 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
| CUST_33815_PI426222305 | JHI_St_60k_v1 | DMT400070920 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
| CUST_33785_PI426222305 | JHI_St_60k_v1 | DMT400070923 | TGCTCAGAAACAGTTTGGAAGAAAATCCGACTCTGCTACAATAACAGCTAAATTTGACCT |
| CUST_33816_PI426222305 | JHI_St_60k_v1 | DMT400070926 | ATAATGGAGGCTGCAGGATTTCATGCTAAGATTGAACTCCAACCTCCTCCTTATCCCATG |
| CUST_33783_PI426222305 | JHI_St_60k_v1 | DMT400070925 | CAGTGAGTAAAACACATGCTGCTCTATTTCTTTTGTTCATGGGATTTCATCTTACATAAC |
Microarray Signals from CUST_33815_PI426222305
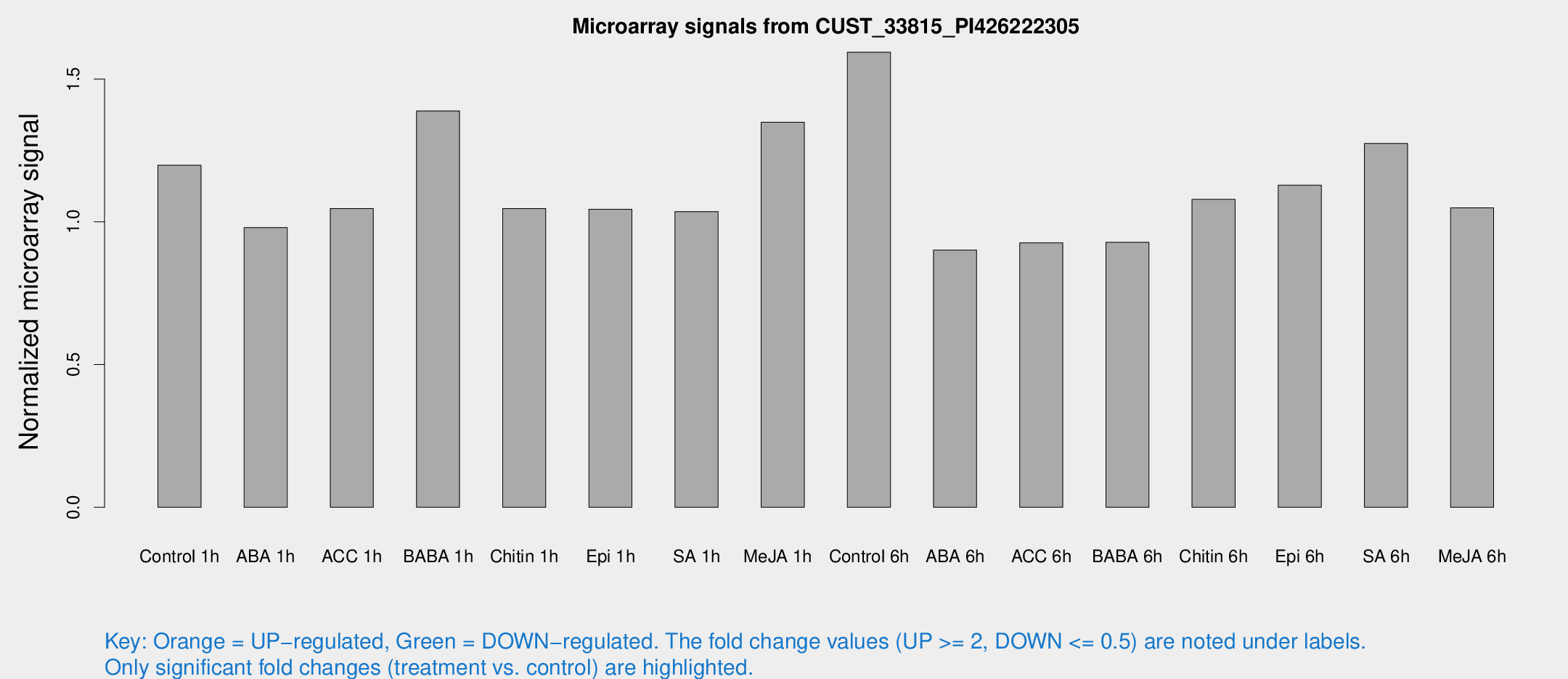
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.7844 | 3.29982 | 1.19845 | 0.539573 |
| ABA 1h | 5.45169 | 3.16236 | 0.979918 | 0.567953 |
| ACC 1h | 6.93633 | 4.13583 | 1.04617 | 0.6067 |
| BABA 1h | 8.85182 | 3.53259 | 1.3884 | 0.619972 |
| Chitin 1h | 5.92206 | 3.26414 | 1.04655 | 0.573934 |
| Epi 1h | 5.70267 | 3.19674 | 1.04408 | 0.585507 |
| SA 1h | 6.90765 | 3.34656 | 1.03592 | 0.530103 |
| Me-JA 1h | 7.09432 | 3.41859 | 1.34898 | 0.682502 |
| Control 6h | 10.2547 | 3.42973 | 1.59402 | 0.561977 |
| ABA 6h | 6.02221 | 3.49245 | 0.900844 | 0.522196 |
| ACC 6h | 6.83722 | 4.05855 | 0.926488 | 0.536682 |
| BABA 6h | 6.54269 | 3.80223 | 0.928299 | 0.538509 |
| Chitin 6h | 7.29942 | 3.77811 | 1.07876 | 0.57134 |
| Epi 6h | 8.11989 | 4.0309 | 1.12858 | 0.572963 |
| SA 6h | 8.30407 | 3.6607 | 1.27421 | 0.623762 |
| Me-JA 6h | 6.65625 | 3.40655 | 1.04923 | 0.551558 |
Source Transcript PGSC0003DMT400070920 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G25100.1 | +2 | 1e-99 | 296 | 153/208 (74%) | Fe superoxide dismutase 1 | chr4:12884649-12886501 REVERSE LENGTH=212 |
