Probe CUST_33785_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33785_PI426222305 | JHI_St_60k_v1 | DMT400070923 | TGCTCAGAAACAGTTTGGAAGAAAATCCGACTCTGCTACAATAACAGCTAAATTTGACCT |
All Microarray Probes Designed to Gene DMG400027577
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33816_PI426222305 | JHI_St_60k_v1 | DMT400070926 | ATAATGGAGGCTGCAGGATTTCATGCTAAGATTGAACTCCAACCTCCTCCTTATCCCATG |
| CUST_33785_PI426222305 | JHI_St_60k_v1 | DMT400070923 | TGCTCAGAAACAGTTTGGAAGAAAATCCGACTCTGCTACAATAACAGCTAAATTTGACCT |
| CUST_33783_PI426222305 | JHI_St_60k_v1 | DMT400070925 | CAGTGAGTAAAACACATGCTGCTCTATTTCTTTTGTTCATGGGATTTCATCTTACATAAC |
| CUST_33815_PI426222305 | JHI_St_60k_v1 | DMT400070920 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
| CUST_33811_PI426222305 | JHI_St_60k_v1 | DMT400070918 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
Microarray Signals from CUST_33785_PI426222305
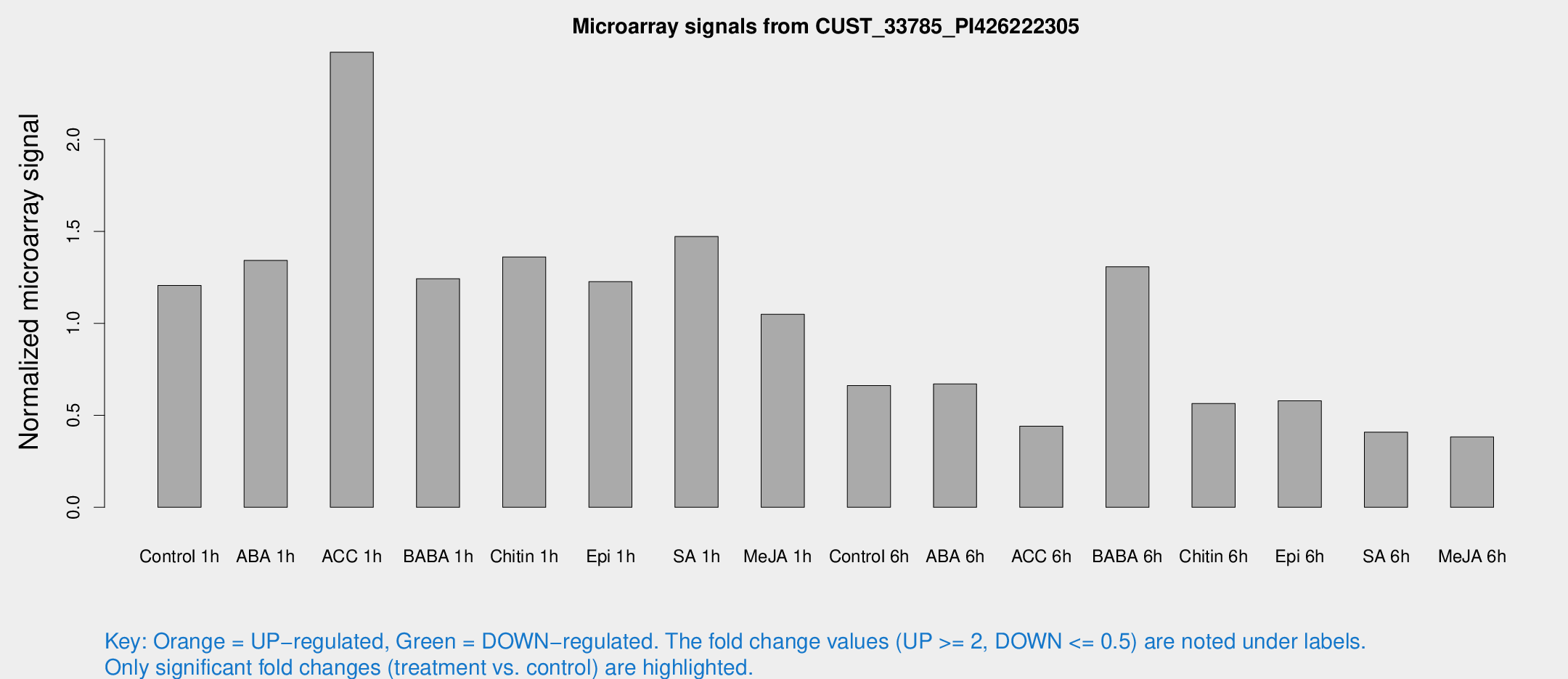
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 155.352 | 32.5263 | 1.20673 | 0.167137 |
| ABA 1h | 147.59 | 12.4166 | 1.34209 | 0.166802 |
| ACC 1h | 362.789 | 143.999 | 2.47454 | 1.30419 |
| BABA 1h | 155.065 | 34.0124 | 1.2432 | 0.18551 |
| Chitin 1h | 163.19 | 40.0727 | 1.36152 | 0.312188 |
| Epi 1h | 131.445 | 8.39027 | 1.22716 | 0.0783103 |
| SA 1h | 218.431 | 87.1171 | 1.47229 | 0.530958 |
| Me-JA 1h | 115.928 | 32.5469 | 1.04992 | 0.295718 |
| Control 6h | 97.3559 | 35.344 | 0.661856 | 0.263432 |
| ABA 6h | 92.2809 | 20.6656 | 0.670574 | 0.10882 |
| ACC 6h | 75.4024 | 33.7144 | 0.441181 | 0.133596 |
| BABA 6h | 182.552 | 17.4205 | 1.30806 | 0.169854 |
| Chitin 6h | 78.2221 | 18.3673 | 0.564821 | 0.118942 |
| Epi 6h | 81.6968 | 9.5446 | 0.578762 | 0.0980945 |
| SA 6h | 53.7189 | 12.8779 | 0.408918 | 0.0696894 |
| Me-JA 6h | 49.2669 | 11.0676 | 0.382703 | 0.057735 |
Source Transcript PGSC0003DMT400070923 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G25100.1 | +2 | 2e-95 | 292 | 149/204 (73%) | Fe superoxide dismutase 1 | chr4:12884649-12886501 REVERSE LENGTH=212 |
