Probe CUST_33816_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33816_PI426222305 | JHI_St_60k_v1 | DMT400070926 | ATAATGGAGGCTGCAGGATTTCATGCTAAGATTGAACTCCAACCTCCTCCTTATCCCATG |
All Microarray Probes Designed to Gene DMG400027577
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33816_PI426222305 | JHI_St_60k_v1 | DMT400070926 | ATAATGGAGGCTGCAGGATTTCATGCTAAGATTGAACTCCAACCTCCTCCTTATCCCATG |
| CUST_33785_PI426222305 | JHI_St_60k_v1 | DMT400070923 | TGCTCAGAAACAGTTTGGAAGAAAATCCGACTCTGCTACAATAACAGCTAAATTTGACCT |
| CUST_33783_PI426222305 | JHI_St_60k_v1 | DMT400070925 | CAGTGAGTAAAACACATGCTGCTCTATTTCTTTTGTTCATGGGATTTCATCTTACATAAC |
| CUST_33815_PI426222305 | JHI_St_60k_v1 | DMT400070920 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
| CUST_33811_PI426222305 | JHI_St_60k_v1 | DMT400070918 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
Microarray Signals from CUST_33816_PI426222305
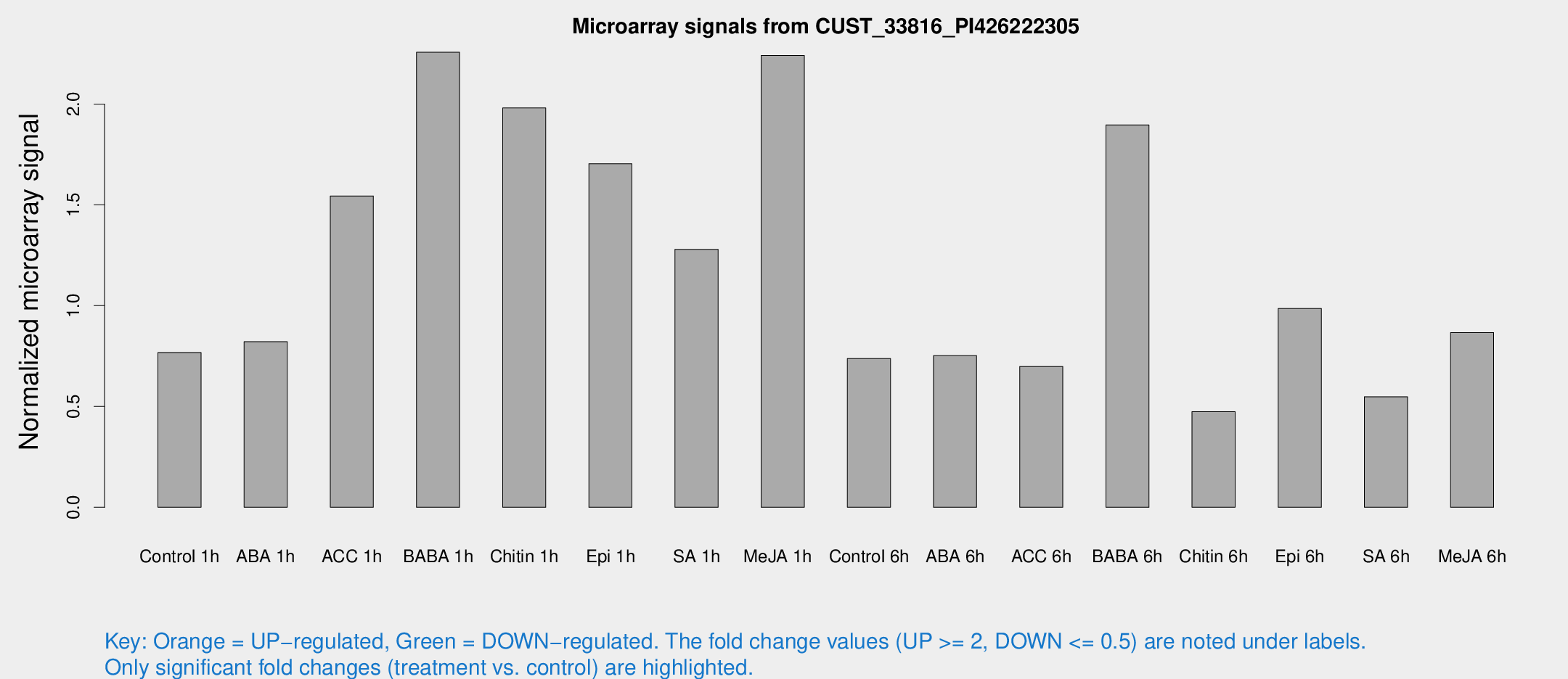
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 16.7977 | 3.72295 | 0.767832 | 0.189049 |
| ABA 1h | 15.9798 | 4.117 | 0.821211 | 0.27324 |
| ACC 1h | 91.9644 | 77.5144 | 1.54343 | 6.54045 |
| BABA 1h | 46.4981 | 7.80243 | 2.25682 | 0.609562 |
| Chitin 1h | 44.0235 | 18.0794 | 1.9804 | 0.75696 |
| Epi 1h | 33.5818 | 9.08191 | 1.70397 | 0.666814 |
| SA 1h | 34.9053 | 18.0641 | 1.27883 | 0.63044 |
| Me-JA 1h | 39.3128 | 7.24844 | 2.24102 | 0.636819 |
| Control 6h | 20.0706 | 10.2576 | 0.738067 | 0.492787 |
| ABA 6h | 24.4809 | 14.9285 | 0.752347 | 0.55502 |
| ACC 6h | 20.9208 | 9.29139 | 0.69851 | 0.329284 |
| BABA 6h | 45.7764 | 9.46161 | 1.89624 | 0.319307 |
| Chitin 6h | 10.7979 | 4.21106 | 0.474484 | 0.200679 |
| Epi 6h | 25.0493 | 7.72176 | 0.985778 | 0.303733 |
| SA 6h | 11.5832 | 4.04012 | 0.548367 | 0.206082 |
| Me-JA 6h | 18.1881 | 3.88474 | 0.866231 | 0.192282 |
Source Transcript PGSC0003DMT400070926 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G25100.1 | +2 | 2e-95 | 289 | 148/199 (74%) | Fe superoxide dismutase 1 | chr4:12884649-12886501 REVERSE LENGTH=212 |
