Probe CUST_33811_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33811_PI426222305 | JHI_St_60k_v1 | DMT400070918 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
All Microarray Probes Designed to Gene DMG400027577
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33816_PI426222305 | JHI_St_60k_v1 | DMT400070926 | ATAATGGAGGCTGCAGGATTTCATGCTAAGATTGAACTCCAACCTCCTCCTTATCCCATG |
| CUST_33785_PI426222305 | JHI_St_60k_v1 | DMT400070923 | TGCTCAGAAACAGTTTGGAAGAAAATCCGACTCTGCTACAATAACAGCTAAATTTGACCT |
| CUST_33783_PI426222305 | JHI_St_60k_v1 | DMT400070925 | CAGTGAGTAAAACACATGCTGCTCTATTTCTTTTGTTCATGGGATTTCATCTTACATAAC |
| CUST_33815_PI426222305 | JHI_St_60k_v1 | DMT400070920 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
| CUST_33811_PI426222305 | JHI_St_60k_v1 | DMT400070918 | CAAAAGAATTACATCAACAAAAGGGCAATGCAATGCTAGTCTAGAAAACTCCACACTCTG |
Microarray Signals from CUST_33811_PI426222305
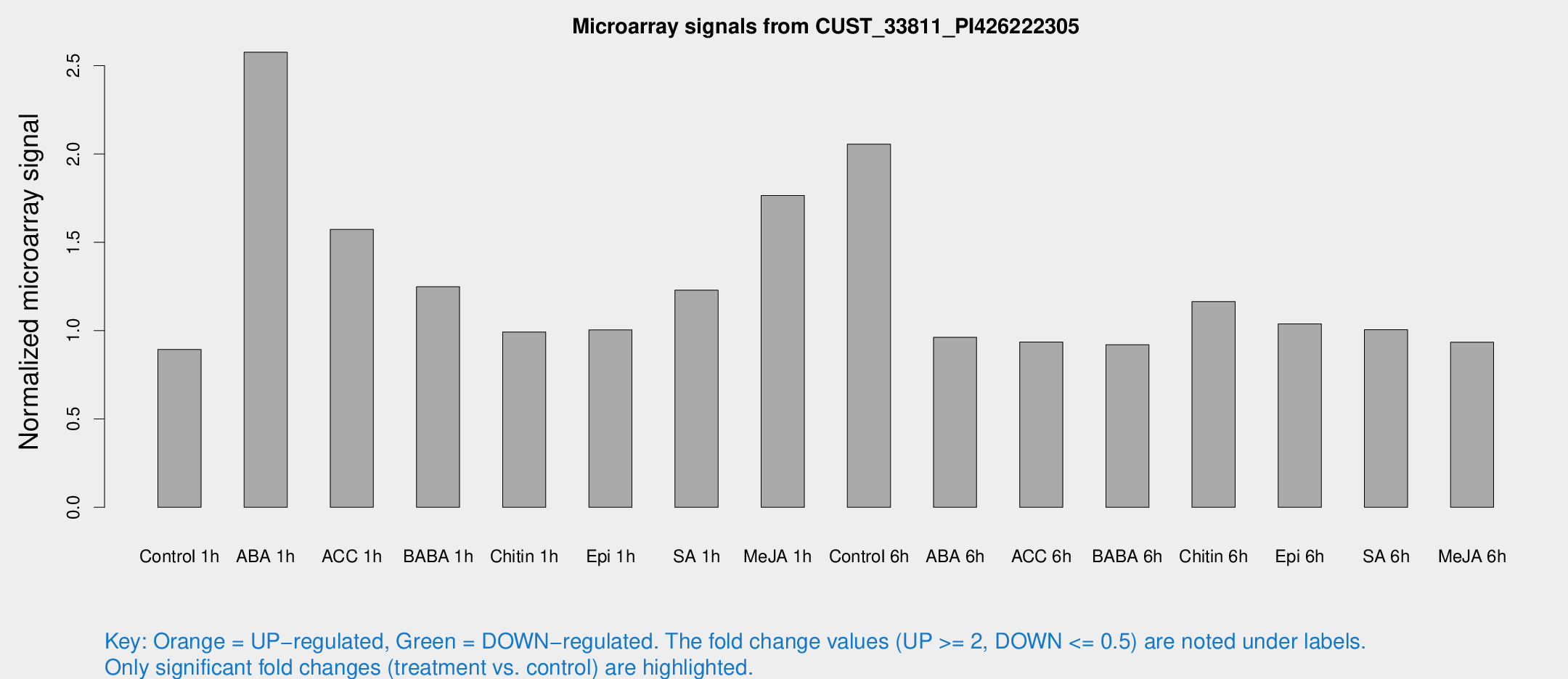
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.89835 | 3.42411 | 0.892947 | 0.517667 |
| ABA 1h | 15.4248 | 3.43343 | 2.57593 | 0.603995 |
| ACC 1h | 11.9869 | 4.41298 | 1.57263 | 0.76303 |
| BABA 1h | 8.35266 | 3.66669 | 1.24864 | 0.610032 |
| Chitin 1h | 5.90492 | 3.40883 | 0.992382 | 0.570255 |
| Epi 1h | 5.7461 | 3.33671 | 1.00393 | 0.581634 |
| SA 1h | 8.68354 | 3.50505 | 1.22882 | 0.546563 |
| Me-JA 1h | 10.4111 | 3.61949 | 1.76508 | 0.721773 |
| Control 6h | 14.2293 | 3.62408 | 2.05552 | 0.596249 |
| ABA 6h | 6.76942 | 3.66307 | 0.961845 | 0.522117 |
| ACC 6h | 7.2487 | 4.30797 | 0.935133 | 0.542169 |
| BABA 6h | 6.81233 | 3.96272 | 0.920382 | 0.534104 |
| Chitin 6h | 8.41296 | 3.95017 | 1.16424 | 0.57784 |
| Epi 6h | 7.96951 | 4.28 | 1.03816 | 0.56483 |
| SA 6h | 6.56253 | 3.80709 | 1.00491 | 0.582563 |
| Me-JA 6h | 6.14264 | 3.56192 | 0.934577 | 0.541729 |
Source Transcript PGSC0003DMT400070918 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G51100.1 | +2 | 2e-96 | 298 | 156/278 (56%) | Fe superoxide dismutase 2 | chr5:20773357-20775635 REVERSE LENGTH=305 |
