Probe CUST_16546_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16546_PI426222305 | JHI_St_60k_v1 | DMT400069190 | GGCTTAAGTTCTCTTTAAAGTTCAAATCATTTTTCGACTTCTACGTTGTCGTCCAAATTG |
All Microarray Probes Designed to Gene DMG400026913
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16819_PI426222305 | JHI_St_60k_v1 | DMT400069191 | GCGGTTGTATAAACTATGCGAAGTTGAATAAAATATTTTGGTCAAAAAATGTCCTTGCCG |
| CUST_16622_PI426222305 | JHI_St_60k_v1 | DMT400069189 | GCAGCTGATTTCTCTACAGATGATATGATTCTTGAAAAGAAACTAGTTCCCCAGAGCTAG |
| CUST_16546_PI426222305 | JHI_St_60k_v1 | DMT400069190 | GGCTTAAGTTCTCTTTAAAGTTCAAATCATTTTTCGACTTCTACGTTGTCGTCCAAATTG |
| CUST_16564_PI426222305 | JHI_St_60k_v1 | DMT400069193 | GGTCCAACTTTAGTCTATCTATGTCTTTCGTTGGTTTTAACATCGAAATTCTACAGCATT |
Microarray Signals from CUST_16546_PI426222305
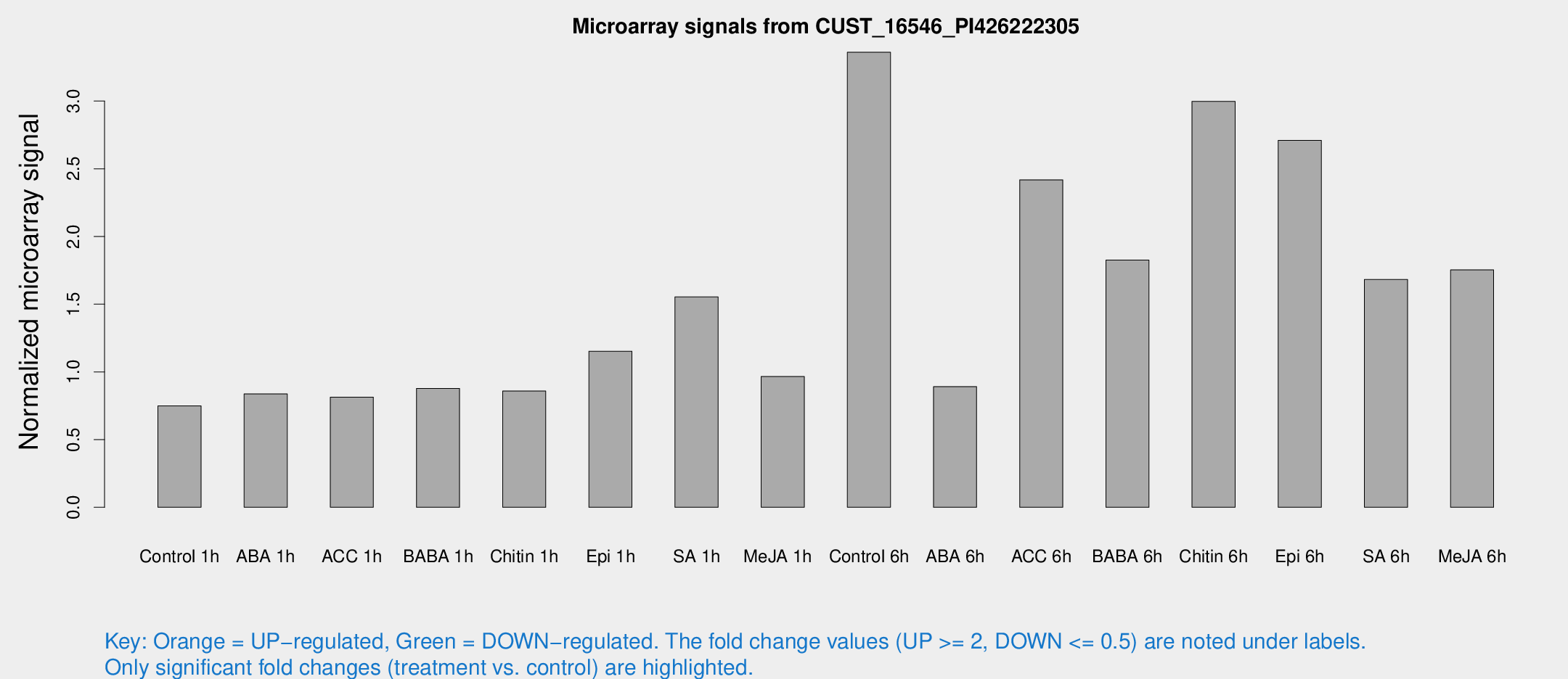
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.94002 | 3.44274 | 0.748793 | 0.433591 |
| ABA 1h | 5.87625 | 3.40885 | 0.83742 | 0.485366 |
| ACC 1h | 6.62587 | 3.84209 | 0.813592 | 0.471504 |
| BABA 1h | 6.72363 | 3.72632 | 0.878155 | 0.486063 |
| Chitin 1h | 6.15899 | 3.59281 | 0.858995 | 0.49799 |
| Epi 1h | 7.96787 | 3.46578 | 1.15293 | 0.513303 |
| SA 1h | 22.0389 | 15.8893 | 1.55383 | 1.74435 |
| Me-JA 1h | 6.28211 | 3.65694 | 0.966122 | 0.559536 |
| Control 6h | 32.0346 | 10.9983 | 3.36093 | 1.42263 |
| ABA 6h | 7.69172 | 3.8721 | 0.891274 | 0.465648 |
| ACC 6h | 24.1799 | 7.46239 | 2.41846 | 0.497221 |
| BABA 6h | 20.3681 | 9.09418 | 1.82556 | 1.01951 |
| Chitin 6h | 34.2739 | 13.64 | 2.99756 | 2.6649 |
| Epi 6h | 30.7819 | 14.202 | 2.71054 | 1.84882 |
| SA 6h | 17.2138 | 9.05836 | 1.68233 | 1.43455 |
| Me-JA 6h | 16.6464 | 6.75009 | 1.75319 | 0.69269 |
Source Transcript PGSC0003DMT400069190 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G09340.1 | +3 | 2e-14 | 75 | 38/46 (83%) | chloroplast RNA binding | chr1:3015473-3018035 FORWARD LENGTH=378 |
