Probe CUST_1251_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1251_PI426222305 | JHI_St_60k_v1 | DMT400001280 | GCAGAAGTGTTCATTTGAACTGCTTTTTCATTTACCTCTTTTTGAACATTTCCAACCTCA |
All Microarray Probes Designed to Gene DMG400000486
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_1100_PI426222305 | JHI_St_60k_v1 | DMT400001281 | CTTGGCCTGAAAATGGCTGCAATCTTACAAACATGTATTCTGTCTTTACAGTATATATCA |
| CUST_863_PI426222305 | JHI_St_60k_v1 | DMT400001279 | ATGATAAGGCTTTGAAGCTTGAAGAGTTTGGAGTGAGTGGCTCAGATGCTGAGAGTGTAT |
| CUST_1251_PI426222305 | JHI_St_60k_v1 | DMT400001280 | GCAGAAGTGTTCATTTGAACTGCTTTTTCATTTACCTCTTTTTGAACATTTCCAACCTCA |
Microarray Signals from CUST_1251_PI426222305
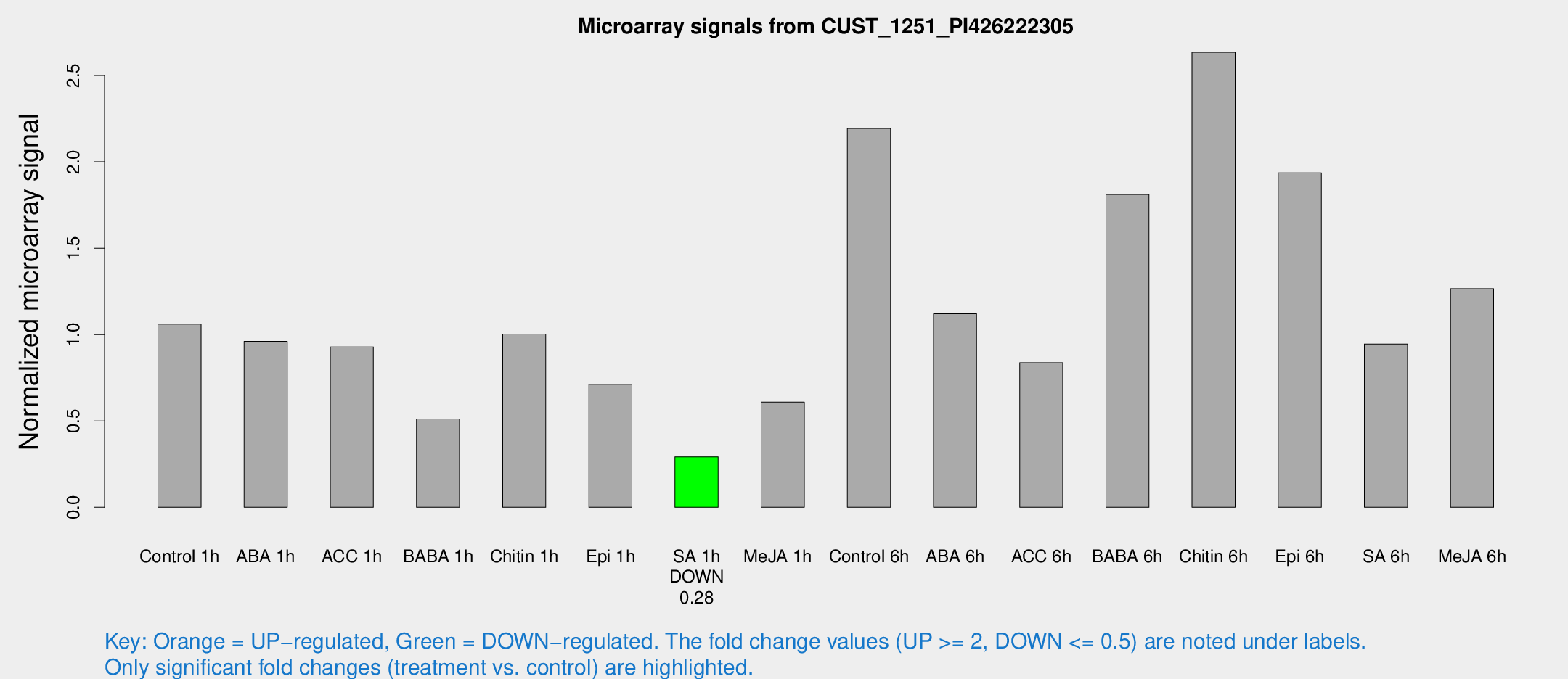
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 19.1075 | 3.30682 | 1.06103 | 0.188952 |
| ABA 1h | 15.1109 | 3.09804 | 0.961047 | 0.203886 |
| ACC 1h | 17.3618 | 3.49651 | 0.928823 | 0.206927 |
| BABA 1h | 10.9074 | 5.48674 | 0.511373 | 0.246788 |
| Chitin 1h | 16.5822 | 3.36255 | 1.00276 | 0.214843 |
| Epi 1h | 11.3278 | 3.11046 | 0.7123 | 0.218956 |
| SA 1h | 5.26772 | 3.05597 | 0.292662 | 0.169621 |
| Me-JA 1h | 9.32564 | 3.21198 | 0.6096 | 0.244008 |
| Control 6h | 41.3048 | 10.1054 | 2.19348 | 0.407367 |
| ABA 6h | 23.9745 | 7.93625 | 1.12055 | 0.420676 |
| ACC 6h | 24.7161 | 13.9603 | 0.837662 | 0.548299 |
| BABA 6h | 42.483 | 15.1541 | 1.81193 | 0.888879 |
| Chitin 6h | 53.1766 | 15.0151 | 2.63467 | 0.734847 |
| Epi 6h | 45.6358 | 19.2127 | 1.93634 | 1.00867 |
| SA 6h | 16.3895 | 3.52498 | 0.945766 | 0.203496 |
| Me-JA 6h | 31.4464 | 14.2467 | 1.26571 | 1.00981 |
Source Transcript PGSC0003DMT400001280 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | Solyc02g086710.2 | +2 | 4e-152 | 439 | 237/241 (98%) | genomic_reference:SL2.50ch02 gene_region:43930486-43935980 transcript_region:SL2.50ch02:43930486..43935980+ go_terms:GO:0005524,GO:0016656 functional_description:Monodehydroascorbate reductase (NADH)-like protein (AHRD V1 **** Q0WUJ1_ARATH); contains Interpro domain(s) IPR013027 FAD-dependent pyridine nucleotide-disulphide oxidoreductase |
| TAIR PP10 | AT3G27820.1 | +2 | 5e-131 | 385 | 204/241 (85%) | monodehydroascorbate reductase 4 | chr3:10315249-10317881 FORWARD LENGTH=488 |
