- Transcript BART1_0-u56729.002
- Transcript IDBART1_0-u56729.002
- Gene IDBART1_0-u56729
Exon Structure of BART1_0-u56729.002
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 640933304 | 640931605 | - |
| chr7H | 2 | 640933929 | 640933717 | - |
| chr7H | 3 | 640934346 | 640934059 | - |
| chr7H | 4 | 640934528 | 640934439 | - |
| chr7H | 5 | 640934999 | 640934818 | - |
| chr7H | 6 | 640936074 | 640935126 | - |
| chr7H | 7 | 640936590 | 640936290 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials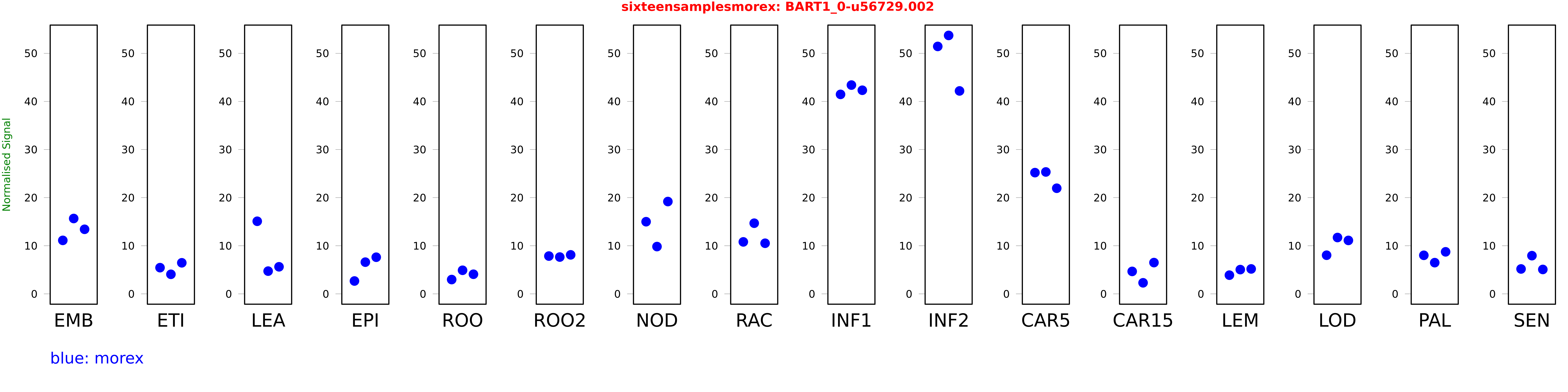
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 11.1189 | 15.6661 | 13.4214 |
| morex | ETI | 5.44387 | 4.06442 | 6.455 |
| morex | LEA | 15.0919 | 4.72576 | 5.62573 |
| morex | EPI | 2.67227 | 6.59067 | 7.61605 |
| morex | ROO | 2.97213 | 4.90149 | 4.07639 |
| morex | ROO2 | 7.85634 | 7.6526 | 8.10868 |
| morex | NOD | 14.9991 | 9.82998 | 19.192 |
| morex | RAC | 10.8034 | 14.6874 | 10.5204 |
| morex | INF1 | 41.4709 | 43.4038 | 42.3292 |
| morex | INF2 | 51.4395 | 53.7267 | 42.1873 |
| morex | CAR5 | 25.2029 | 25.3437 | 21.9575 |
| morex | CAR15 | 4.6672 | 2.29721 | 6.4991 |
| morex | LEM | 3.88507 | 5.04248 | 5.18336 |
| morex | LOD | 8.03119 | 11.7121 | 11.1103 |
| morex | PAL | 8.01423 | 6.48716 | 8.73679 |
| morex | SEN | 5.17668 | 7.93898 | 5.07412 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g44230.1 | +2 | 0.0 | 874 | 446/614 (73%) | protein|transcription factor X1, putative, expressed |
| TAIR PP10 | AT3G48670 @ ARAPORT AT3G48670.2 @ TAIR |
+2 | 2e-135 | 427 | 245/621 (39%) | Symbols: IDN2, RDM12 | XH/XS domain-containing protein | chr3:18031240-18033615 FORWARD LENGTH=647 |
| BRACH PP3 | Bradi2g44450.2.p | +2 | 0.0 | 952 | 496/614 (81%) | pacid=32771748 transcript=Bradi2g44450.2 locus=Bradi2g44450 ID=Bradi2g44450.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3723 bp)
>BART1_0-u56729.002 3723 150831_barley_pseudomolecules CCGATACTAACGTGCAATGGTGTAACGGAAAACGTACCCGCAGCCAATCTCCCGGCAGCA TCTGATCCCTCCATTCTCTATCCGGCGGCTCAGATCCATGGCTGCATATGCATTCCGGCA TCACCTCTCCGGTTCGAGCAGGAGCAGGTAATCCCAGTCCCCTCCCGAGCTCCACCACCA TTCCCCTCCCCCCTCACTCCCGCAGGACCCTAGCCGCCGCCGCCGCCGCCGCCCCCCACC AGCCGGGCTTCTTCTCCTCCTCCCCGACCCCGGGTTCCTTCTCTGCCTCGGCGACCCAAA GTCACTGTTTCCGGCAGGTTGTCAAAAAAAAAAAAAATCCTTCTCGGAGATGGAATACAG CTCAGACGACGATTCGGATATCAGCGACTCCGAGATCGACGAGTACGAAGGCAAGATTAA CGCCCGTCTGATTTCAGGAGACTTAAAGTTCAGGAATGGCGATAGCTACGGTTGCCCATT CTGCACTGGCAAGAAGAACAAAGGTTACAACATGCAGAGCCTTCTTCAGCATTCCTCGGG GGTAGGCGCAGCGCCTAATCGGCCGGCAAAAGATAAAGCGTCGCACCGCGCCCTCGCCAA GCATTTGAAGAATGACGTTGCTAAACCTTCTGAGCCACAGCAACAGCAGCCGCAGCAGAT TGTTGTGGAGCCGCAGCCTCTTCCAAACAGACATGAGAAGTTTGTGTGGCCCTGGATGGG TGTTCTAGTTAATGTACCTACTGAATGGAAGGACGGGCGTCAGGTTGGAGAAAGTGGTAC TCGTTTGAAGGGGGAGCTGTCGCAATTTTGCCCGCTGAAGGTTATTCCTCTATGGAATTT CCGAGGCCATACAGGGAATGCTATTGTTGAGTTTGCAAAAGACTGGAATGGTTTCAGAAA TGCACTTGCATTTGAACAGTACTTTGAGGCAGGCGGCTGTGGCAGAAGGGACTGGAAGCA AAACCAGAATCAGGGGTCAAAGCTTTTTGGATGGGTTGCAAGGGCTGAAGACTACAATCT TTCGGGGCTAATCGGGGACCACTTAAGGAAAAATGCTGACTTGAAAACTATCGATGACCT TGAGAATGAAGGAACACGCAAAAATAATAAACTTGTAGCTAATTTGGCTAACCAAATTGA GGTCAAGAATAAGTATTTAGAGGAGCTTGAACTTAGATACAATGAGACAACTGCGTCACT TGAGAAGATGATGGGGCAAAGGGAACAACGTCTCCAAGCGTATAATGAAGAAATTCGGAA GATGCAGCAGCTAGCTCGCAGACATTCTGAAAGAATTATTGATGAGAACCAAAATCTGCG TTCGGAACTGGAGTCCAAGATGAGTGAACTTAATGAAAGATCCAAAGAGCTTGATGACCT TTCTGCAAAAAGTTCTCATGACAGAAGTAATCTTGAGCAGGAGAAGCAGAAGAATGCTAT TAAATCAAACCATCTTAAGTTGGCAACAGTGGAACAACAGAGAGCTAATGAGGATGTGGT GAAGCTTGTGAGAGATCAGAAGAGAGAGAAAGTTGCTGCATTAAACAAGATCCTGGAGTT AGAACAGCAGTTGGAGGCAAAACAGACGCTTGAACTGGAAATACAGCAGCTGAAGGGCAA ATTGGAAGTGATGAAGCATATGCCAGGTCATGAAGATTCAGAGGCGAAGGATAAAATCAA TCAACTTAGTGAGGAGCTGCAAGATAAGATGGATGAACTGGATGCTATGGAGTCACTTAA CCAAACTCTGGTTATTAAAGAAAGCAAAAGCAATACTGAGATGCAAGAAGCTCGGAAGGA GCTGGAAAATGGCTTGCTTAATCTTCCAGGCGGCGGAGCACATATAGGAATTAAGAGAAT GGGCGAGCTTGATCTGAAAGCATTTTCAAATGCTCTCGGACAGAAGTTGTCGAAAGAGGA AGCAGAAGTTACTGCTGCCATTCTTTGTTCAAAGTGGGAAGCTGAAATTAGGAATCCAGA ATGGCACCCTTTTAGGACTGTCATGGTTGACGGAAAGGAAGTGGAGAAAATCGATGCTGA TGACACTAAGCTTCAAGAATTAAAAGATGAGCATGGCCAAGAAATCTATTCGTTGGTGAC CAAGGCGCTGCGTGAGTACAACGCCAATAGCACCAGGTATCCCGTAGGAGAGCTGTGGAA CTTCAGAGAAGAACGGAAGGCGTCTCTCAAGGAAGCCGTTCAGGTGGTCCTGAGGCAGTG GCGAACCACCAGGAGGAAGCGTTGATTGAGCCCAACCAATGTCGATTCTTTCCTCCTCGT ATGGACATCGACATTAGCGGTGTGGTTTGATAAAATCTCTAGTGGCTATGTACGCCCATG TTGGATATAAGTTGAATAATAAATAATCCTGTTGTTTCAGTAGTAGTAGCATTAGTGAAG TATCTATCCATCTGTGTAAACAAGCTTGAATAAAGACTTGGAGTAGTATCAGCACCTGCA AGCACGCTATCAACCCGATCATCGGTGTCTGTTGGTAAGCGCCGCTGTATGCTTTGTTTC TGAGTTGCTGATAGTAATTCAGAGGAGTGTTTGTTTGTTGCCTGAATTAATGGTGACCTC CTGTACCAAGTATAGATACAGGAATGCATGAATTAGCACATCCACCTAAATACAATGATG CAAGTTACTGTGAGGCAACATTGATCTCCAATGAATAGAGAGGTACTATTTGAGAGTAAA AAAAAATCATTCTATGCCAAGGGGAAAAGGTACGGGGTAACATTTGTAAAAGATGGATTA CCTCCCTATTACATCACCAGCCATATTAATCTGAAAGCGTAGACATCTCATTCAGCATCT CAAATAATGTTGGGAGTACACAAGCAGGTAAGCAACTTGTAATACCTGAAGATAAACAAT ATTAAAGTAGCTGTTTGCCTTCACTAACAGCTACTGATTCAAACCTCAGTCAACATATGT ATTTTGGGTTTGGATAAAGTTAATACTAGTTCAACAACCTTTATACGAATAAAATTGGCA ACAGAGGACCCCCAAAGAATAGTTGCTTTGTTGCATCTATATCCATATGGACCAATATCA TCCGCACAGGATGCACAACTACAACAGCTGAATATTGACAAAGTTATACAGCTACTTTTT TTACAACAATAGGCCGAATAGTTACAGCGGGCCCTCGATGACTAACCTACAGCTAGTGTA CACTCCGACTCCCGATGCCTGGTAGCAACAACCAGGCAAAACAGCTGGGACTGGGATCAC ATGTGCCTCCATGCGATCGCCGGAACCTAAACCACATGTACAGGTGCGCTTTCCCCAACC TCGCTACCGATCAGCAGGTACAGCAATCTATGTTAGGACACGGCGTCGCCATATCTTTGT TATTGTGCTAGCTCTCCATTGCGCTCTCGGCGTCGTCAAGCACGTCTGCGCTTTCATCAA GAAGGTGGTCGCACAGCTCCAGGAGCTCGTCGGCCAGTGGAGGGATGTGAGGATGCCGCG TCTCGTTGTACATGTGCGCATTGTCTGCTATCTGCGCCACGTCATTCCTGAATTCGTACC TGTTTTTGTACTCCATCTTCCTCGCCTTGTCCCTGATGGTACCCAGATCCATTGGGCGCT GGATTACATCGTAGTAATCGGGAGCGACTTTCTTTGTCACTGGTTTAAGAAACAGCAATG ATATAGCAGTTTGGTTCCGCAAGTGGTCAACTATCTTCTCCAATATGTTGGAGAGCTCAA CCT
Protein Sequence (631 aa)
>BART1_0-u56729.002 631 150831_barley_pseudomolecules MEYSSDDDSDISDSEIDEYEGKINARLISGDLKFRNGDSYGCPFCTGKKNKGYNMQSLLQ HSSGVGAAPNRPAKDKASHRALAKHLKNDVAKPSEPQQQQPQQIVVEPQPLPNRHEKFVW PWMGVLVNVPTEWKDGRQVGESGTRLKGELSQFCPLKVIPLWNFRGHTGNAIVEFAKDWN GFRNALAFEQYFEAGGCGRRDWKQNQNQGSKLFGWVARAEDYNLSGLIGDHLRKNADLKT IDDLENEGTRKNNKLVANLANQIEVKNKYLEELELRYNETTASLEKMMGQREQRLQAYNE EIRKMQQLARRHSERIIDENQNLRSELESKMSELNERSKELDDLSAKSSHDRSNLEQEKQ KNAIKSNHLKLATVEQQRANEDVVKLVRDQKREKVAALNKILELEQQLEAKQTLELEIQQ LKGKLEVMKHMPGHEDSEAKDKINQLSEELQDKMDELDAMESLNQTLVIKESKSNTEMQE ARKELENGLLNLPGGGAHIGIKRMGELDLKAFSNALGQKLSKEEAEVTAAILCSKWEAEI RNPEWHPFRTVMVDGKEVEKIDADDTKLQELKDEHGQEIYSLVTKALREYNANSTRYPVG ELWNFREERKASLKEAVQVVLRQWRTTRRKR
