- Transcript BART1_0-u56729.001
- Transcript IDBART1_0-u56729.001
- Gene IDBART1_0-u56729
Exon Structure of BART1_0-u56729.001
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 640933304 | 640931603 | - |
| chr7H | 2 | 640933929 | 640933717 | - |
| chr7H | 3 | 640934346 | 640934059 | - |
| chr7H | 4 | 640934528 | 640934439 | - |
| chr7H | 5 | 640934999 | 640934818 | - |
| chr7H | 6 | 640936058 | 640935126 | - |
| chr7H | 7 | 640936590 | 640936290 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials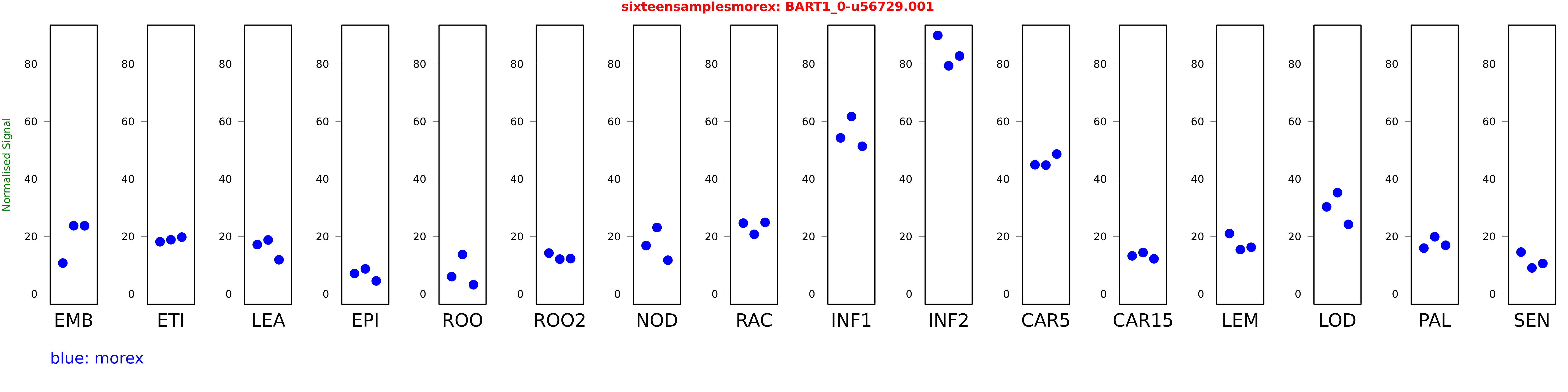
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 10.7085 | 23.7081 | 23.6904 |
| morex | ETI | 18.1367 | 18.8396 | 19.7418 |
| morex | LEA | 17.1504 | 18.7526 | 11.8647 |
| morex | EPI | 7.06199 | 8.69231 | 4.51039 |
| morex | ROO | 5.97982 | 13.6835 | 3.16497 |
| morex | ROO2 | 14.1832 | 12.093 | 12.2538 |
| morex | NOD | 16.8105 | 23.0905 | 11.718 |
| morex | RAC | 24.621 | 20.7191 | 24.8877 |
| morex | INF1 | 54.2815 | 61.7266 | 51.3714 |
| morex | INF2 | 89.9448 | 79.3722 | 82.78 |
| morex | CAR5 | 44.9281 | 44.8091 | 48.6542 |
| morex | CAR15 | 13.2226 | 14.3835 | 12.1926 |
| morex | LEM | 20.9756 | 15.3997 | 16.224 |
| morex | LOD | 30.2816 | 35.2104 | 24.1902 |
| morex | PAL | 15.9086 | 19.8704 | 16.9191 |
| morex | SEN | 14.5321 | 9.02586 | 10.5682 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g44230.1 | +1 | 0.0 | 873 | 446/614 (73%) | protein|transcription factor X1, putative, expressed |
| TAIR PP10 | AT3G48670 @ ARAPORT AT3G48670.2 @ TAIR |
+1 | 3e-135 | 427 | 245/621 (39%) | Symbols: IDN2, RDM12 | XH/XS domain-containing protein | chr3:18031240-18033615 FORWARD LENGTH=647 |
| BRACH PP3 | Bradi2g44450.2.p | +1 | 0.0 | 949 | 496/614 (81%) | pacid=32771748 transcript=Bradi2g44450.2 locus=Bradi2g44450 ID=Bradi2g44450.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3709 bp)
>BART1_0-u56729.001 3709 150831_barley_pseudomolecules CCGATACTAACGTGCAATGGTGTAACGGAAAACGTACCCGCAGCCAATCTCCCGGCAGCA TCTGATCCCTCCATTCTCTATCCGGCGGCTCAGATCCATGGCTGCATATGCATTCCGGCA TCACCTCTCCGGTTCGAGCAGGAGCAGGTAATCCCAGTCCCCTCCCGAGCTCCACCACCA TTCCCCTCCCCCCTCACTCCCGCAGGACCCTAGCCGCCGCCGCCGCCGCCGCCCCCCACC AGCCGGGCTTCTTCTCCTCCTCCCCGACCCCGGGTTCCTTCTCTGCCTCGGCGACCCAAA GGTTGTCAAAAAAAAAAAAAATCCTTCTCGGAGATGGAATACAGCTCAGACGACGATTCG GATATCAGCGACTCCGAGATCGACGAGTACGAAGGCAAGATTAACGCCCGTCTGATTTCA GGAGACTTAAAGTTCAGGAATGGCGATAGCTACGGTTGCCCATTCTGCACTGGCAAGAAG AACAAAGGTTACAACATGCAGAGCCTTCTTCAGCATTCCTCGGGGGTAGGCGCAGCGCCT AATCGGCCGGCAAAAGATAAAGCGTCGCACCGCGCCCTCGCCAAGCATTTGAAGAATGAC GTTGCTAAACCTTCTGAGCCACAGCAACAGCAGCCGCAGCAGATTGTTGTGGAGCCGCAG CCTCTTCCAAACAGACATGAGAAGTTTGTGTGGCCCTGGATGGGTGTTCTAGTTAATGTA CCTACTGAATGGAAGGACGGGCGTCAGGTTGGAGAAAGTGGTACTCGTTTGAAGGGGGAG CTGTCGCAATTTTGCCCGCTGAAGGTTATTCCTCTATGGAATTTCCGAGGCCATACAGGG AATGCTATTGTTGAGTTTGCAAAAGACTGGAATGGTTTCAGAAATGCACTTGCATTTGAA CAGTACTTTGAGGCAGGCGGCTGTGGCAGAAGGGACTGGAAGCAAAACCAGAATCAGGGG TCAAAGCTTTTTGGATGGGTTGCAAGGGCTGAAGACTACAATCTTTCGGGGCTAATCGGG GACCACTTAAGGAAAAATGCTGACTTGAAAACTATCGATGACCTTGAGAATGAAGGAACA CGCAAAAATAATAAACTTGTAGCTAATTTGGCTAACCAAATTGAGGTCAAGAATAAGTAT TTAGAGGAGCTTGAACTTAGATACAATGAGACAACTGCGTCACTTGAGAAGATGATGGGG CAAAGGGAACAACGTCTCCAAGCGTATAATGAAGAAATTCGGAAGATGCAGCAGCTAGCT CGCAGACATTCTGAAAGAATTATTGATGAGAACCAAAATCTGCGTTCGGAACTGGAGTCC AAGATGAGTGAACTTAATGAAAGATCCAAAGAGCTTGATGACCTTTCTGCAAAAAGTTCT CATGACAGAAGTAATCTTGAGCAGGAGAAGCAGAAGAATGCTATTAAATCAAACCATCTT AAGTTGGCAACAGTGGAACAACAGAGAGCTAATGAGGATGTGGTGAAGCTTGTGAGAGAT CAGAAGAGAGAGAAAGTTGCTGCATTAAACAAGATCCTGGAGTTAGAACAGCAGTTGGAG GCAAAACAGACGCTTGAACTGGAAATACAGCAGCTGAAGGGCAAATTGGAAGTGATGAAG CATATGCCAGGTCATGAAGATTCAGAGGCGAAGGATAAAATCAATCAACTTAGTGAGGAG CTGCAAGATAAGATGGATGAACTGGATGCTATGGAGTCACTTAACCAAACTCTGGTTATT AAAGAAAGCAAAAGCAATACTGAGATGCAAGAAGCTCGGAAGGAGCTGGAAAATGGCTTG CTTAATCTTCCAGGCGGCGGAGCACATATAGGAATTAAGAGAATGGGCGAGCTTGATCTG AAAGCATTTTCAAATGCTCTCGGACAGAAGTTGTCGAAAGAGGAAGCAGAAGTTACTGCT GCCATTCTTTGTTCAAAGTGGGAAGCTGAAATTAGGAATCCAGAATGGCACCCTTTTAGG ACTGTCATGGTTGACGGAAAGGAAGTGGAGAAAATCGATGCTGATGACACTAAGCTTCAA GAATTAAAAGATGAGCATGGCCAAGAAATCTATTCGTTGGTGACCAAGGCGCTGCGTGAG TACAACGCCAATAGCACCAGGTATCCCGTAGGAGAGCTGTGGAACTTCAGAGAAGAACGG AAGGCGTCTCTCAAGGAAGCCGTTCAGGTGGTCCTGAGGCAGTGGCGAACCACCAGGAGG AAGCGTTGATTGAGCCCAACCAATGTCGATTCTTTCCTCCTCGTATGGACATCGACATTA GCGGTGTGGTTTGATAAAATCTCTAGTGGCTATGTACGCCCATGTTGGATATAAGTTGAA TAATAAATAATCCTGTTGTTTCAGTAGTAGTAGCATTAGTGAAGTATCTATCCATCTGTG TAAACAAGCTTGAATAAAGACTTGGAGTAGTATCAGCACCTGCAAGCACGCTATCAACCC GATCATCGGTGTCTGTTGGTAAGCGCCGCTGTATGCTTTGTTTCTGAGTTGCTGATAGTA ATTCAGAGGAGTGTTTGTTTGTTGCCTGAATTAATGGTGACCTCCTGTACCAAGTATAGA TACAGGAATGCATGAATTAGCACATCCACCTAAATACAATGATGCAAGTTACTGTGAGGC AACATTGATCTCCAATGAATAGAGAGGTACTATTTGAGAGTAAAAAAAAATCATTCTATG CCAAGGGGAAAAGGTACGGGGTAACATTTGTAAAAGATGGATTACCTCCCTATTACATCA CCAGCCATATTAATCTGAAAGCGTAGACATCTCATTCAGCATCTCAAATAATGTTGGGAG TACACAAGCAGGTAAGCAACTTGTAATACCTGAAGATAAACAATATTAAAGTAGCTGTTT GCCTTCACTAACAGCTACTGATTCAAACCTCAGTCAACATATGTATTTTGGGTTTGGATA AAGTTAATACTAGTTCAACAACCTTTATACGAATAAAATTGGCAACAGAGGACCCCCAAA GAATAGTTGCTTTGTTGCATCTATATCCATATGGACCAATATCATCCGCACAGGATGCAC AACTACAACAGCTGAATATTGACAAAGTTATACAGCTACTTTTTTTACAACAATAGGCCG AATAGTTACAGCGGGCCCTCGATGACTAACCTACAGCTAGTGTACACTCCGACTCCCGAT GCCTGGTAGCAACAACCAGGCAAAACAGCTGGGACTGGGATCACATGTGCCTCCATGCGA TCGCCGGAACCTAAACCACATGTACAGGTGCGCTTTCCCCAACCTCGCTACCGATCAGCA GGTACAGCAATCTATGTTAGGACACGGCGTCGCCATATCTTTGTTATTGTGCTAGCTCTC CATTGCGCTCTCGGCGTCGTCAAGCACGTCTGCGCTTTCATCAAGAAGGTGGTCGCACAG CTCCAGGAGCTCGTCGGCCAGTGGAGGGATGTGAGGATGCCGCGTCTCGTTGTACATGTG CGCATTGTCTGCTATCTGCGCCACGTCATTCCTGAATTCGTACCTGTTTTTGTACTCCAT CTTCCTCGCCTTGTCCCTGATGGTACCCAGATCCATTGGGCGCTGGATTACATCGTAGTA ATCGGGAGCGACTTTCTTTGTCACTGGTTTAAGAAACAGCAATGATATAGCAGTTTGGTT CCGCAAGTGGTCAACTATCTTCTCCAATATGTTGGAGAGCTCAACCTGC
Protein Sequence (631 aa)
>BART1_0-u56729.001 631 150831_barley_pseudomolecules MEYSSDDDSDISDSEIDEYEGKINARLISGDLKFRNGDSYGCPFCTGKKNKGYNMQSLLQ HSSGVGAAPNRPAKDKASHRALAKHLKNDVAKPSEPQQQQPQQIVVEPQPLPNRHEKFVW PWMGVLVNVPTEWKDGRQVGESGTRLKGELSQFCPLKVIPLWNFRGHTGNAIVEFAKDWN GFRNALAFEQYFEAGGCGRRDWKQNQNQGSKLFGWVARAEDYNLSGLIGDHLRKNADLKT IDDLENEGTRKNNKLVANLANQIEVKNKYLEELELRYNETTASLEKMMGQREQRLQAYNE EIRKMQQLARRHSERIIDENQNLRSELESKMSELNERSKELDDLSAKSSHDRSNLEQEKQ KNAIKSNHLKLATVEQQRANEDVVKLVRDQKREKVAALNKILELEQQLEAKQTLELEIQQ LKGKLEVMKHMPGHEDSEAKDKINQLSEELQDKMDELDAMESLNQTLVIKESKSNTEMQE ARKELENGLLNLPGGGAHIGIKRMGELDLKAFSNALGQKLSKEEAEVTAAILCSKWEAEI RNPEWHPFRTVMVDGKEVEKIDADDTKLQELKDEHGQEIYSLVTKALREYNANSTRYPVG ELWNFREERKASLKEAVQVVLRQWRTTRRKR
