- Transcript BART1_0-u56048.003
- Transcript IDBART1_0-u56048.003
- Gene IDBART1_0-u56048
Exon Structure of BART1_0-u56048.003
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 620784789 | 620784689 | + |
| chr7H | 2 | 620785405 | 620785279 | + |
| chr7H | 3 | 620786382 | 620785866 | + |
| chr7H | 4 | 620786624 | 620786494 | + |
| chr7H | 5 | 620787133 | 620786727 | + |
| chr7H | 6 | 620788394 | 620787261 | + |
| chr7H | 7 | 620788805 | 620788653 | + |
| chr7H | 8 | 620789899 | 620788920 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials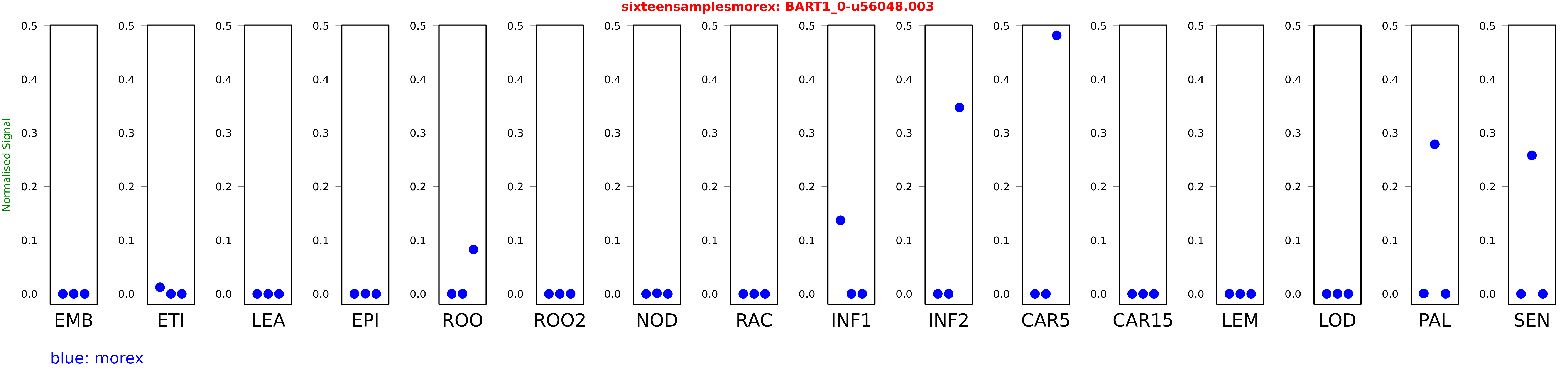
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0.0122965 | 0.00000000410064 | 0.0000000671875 |
| morex | LEA | 0.00000164078 | 0 | 0 |
| morex | EPI | 0 | 0.000256155 | 0 |
| morex | ROO | 0 | 0 | 0.0827132 |
| morex | ROO2 | 0 | 0.0000177423 | 0 |
| morex | NOD | 0 | 0.00118176 | 0 |
| morex | RAC | 0.000000017767 | 0 | 0 |
| morex | INF1 | 0.137265 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0.347491 |
| morex | CAR5 | 0 | 0 | 0.481875 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0 | 0.0000144885 | 0.000000000533225 |
| morex | PAL | 0.000675988 | 0.278947 | 0 |
| morex | SEN | 0 | 0.258117 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os06g46240.1 | +1 | 0.0 | 660 | 344/514 (67%) | protein|BTB/POZ domain containing protein, putative, expressed |
| TAIR PP10 | AT1G04390 @ ARAPORT AT1G04390.1 @ TAIR |
+1 | 4e-69 | 249 | 145/362 (40%) | Symbols: | BTB/POZ domain-containing protein | chr1:1179678-1183615 REVERSE LENGTH=849 |
| BRACH PP3 | Bradi1g32690.2.p | +1 | 0.0 | 1512 | 780/992 (79%) | pacid=32804374 transcript=Bradi1g32690.2 locus=Bradi1g32690 ID=Bradi1g32690.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3550 bp)
>BART1_0-u56048.003 3550 150831_barley_pseudomolecules NNNNNNNGCGCCGGCGCCGGCGCCGGGGAGCATCTCCTCACCGACCGCGTCCTCTCCCTC CGCGCACGCCTGCACGACGCCCTCTCCCTCGGCCTCGCCAGGTCTGATGGCCACGGTGCA AAAAAATGGCAGTCTACTGATGCTGGAATTCAGTCTCACGCGATCAAAGCAGTGACTGCT TTTCTCGGTTGTTTGTCAACTGAAATGCTGCGGCTTCCTCCTATAAAGGAGTCAATTTCG GATATTCTCGTACTCTTGGAAGGTGTTCTTCAGACAAAGAACGTTTCAGTTCTGATCCAA GCAGCAGATGTGAGCTCAAAGTTAGTTTCCAGCATAGGCAATTCGATTCGGCAATACTCA ATTTTGGAGATTGTTTCGCCCCTCTCATGCCAGTTGTCGGCACACCAGTTACCTACGGCT ATCTCATGTGCAAGGGCAATAAACTGCATATTGAATAGCCTAGTGACAGCGAGAGGTTCA ACTCATGCAGAAATTTTGGAAACCTTGGAGAGAACCAATGCAGTTGCTAGTATTGTTTCA GCTCTGCAGAGTTACACTCTTGATGTCCATCCACTGAGTTATCTGACCGAGATGATATCA CTGCTAAGAACCATACTGTGGACTTGGCCTTCCTCAAGATACCATGTATGGAGTAACGGC GACCTGATGGGGAAGTTAGCACATTACTGTCTTGCCACTGAAACAACGGTTGTAGCTAAA GTTCTGAGGCTGTATGCTGCTTTAGCTTTATGTGGCCATGGAGCTACGATCCTTCTCAAG AATGAAGTCTTAATGGCTAAGATTTGTGAGCTTGTGGGGACGTCCCATCCGACTAGTACT AGGATTGAAGCGTTCAAACTCTGCCATGTTCTTTTGAGATCTTCGAAGGGCTGTAGTCAG TTAATGACTTCACGTTGCCAGCCTATTGTTGAAGGCATAATCGATGCAATGCGTGCAAAG GATGATAAATTGTTAGTAACAGAGGGGTGTCGTACTGCACTGGTGGTCCTCCGTTATGCT GGGAATCATCATCAATGCTTTTGGTCTAATGCCATTGATAAAGTATTATATACCATTCTT ACGGGCCGCTGCCTCTCCTCGCATCAAAGTGATCAAGTTTTGTGCGATGATGAGCTATTT GATATGGTTTCCAAAAACTTCATGGATATACATCCATACGTCTGGGATATACTTGGATAT CTAGTGGTGCATTGCACTGACAAGCACCATCCTGTAAGGAAAGGGAAAGGTCATAGCTTG CACGCACTGATATCTTGTGCTTGCTTGCTGGCCACAGATGTTATGCGCAAAAGCAGCCCC ATGAAATTGTCCAAAGATGTGCAGGAGCCGGCTTTGAGAGCAGTTCTGATGATGCTTCTC TCTCCCAATGGATATATTCAGTATGAGGCAAGTTTTAAACTTTCAGAAGGTCTTCCGTAT TTGGGTGATGGCTATTTGAACGTTCTATTGTCATCATTAGAATCAAATGCTACGAGAAGT GTTGCCACATCTTTTGACAGTTTCAAAATTATGACCAACCTCATGAACCTAGCATGTTTG GTGCTGTCACGACCTTACCATAATCTCCTGAACAAGAGGAATCCTATAGATGTGTTGTCC ACCATTATCAAGGAATCTCTCCACAATCATATACATATTACGAGGTCAAAAGTCACTTCC CATCTGCATTTTTGTTTTGATGGAAGTTCATGCTGTTGCTATCTCGGTGAAGAGTGGGAG GGTGAAAACATAGTTTTATTCTACGGTCTTGTTGTGCTGTTTAATCTTATGAGGAGCACT ACTTTAGTTTGTGTTCAGTGTAAGAGGAAGTTGGATGTTGGAAATCTGTGCCATGACTGC AGAGATCAATATACAGAAGATTTTCTTATAGTTCTTCAACATGCTCTAAGTCAAAGCTTG AGTTCTGGACCGAAGCTCTATATTGCACACATCCTGAGTTTGTTTGGCCTCTGTGGTTCT CCAAGCAAGTTGGGCGGGAAGATGACTAGTGCATTAGATAATAACGATCTAGCTGATCTG GAACTTTTGCTTTCAGATGGTGAATCTCTAAATGCTCACACAGCCATCATATCAGCGAGG TGTCCTAAGCTGTTGCCATCTGTAAAATATCTACTTGGCAGCGATGAAAAAGTTAAAGAT GAATGGGGAAAATCAGTGTACCGAGTTCAGATGTCTGATCGTGTAGATAGTCGCGCCTTG AAGAAGATTTTGGAGTACACATATACTGGTTTTCTGATGGTAGGCGATGACGTTGTTAAA CCAGTAAGGACACTTGCGAAGTATTGTCACCTGAAACCATTGGCAGAAATGCTTCAGAAA GAGCAACCCAGATGGAACTCCGATTATGCCAGATATGATCTTAGTGCTGCACTTGGACCT GTTGAGCATTCATTCTCGGATATAATTTTGGAGGCTAGGTCAAATGAGGAAATGAATTGT CACCATGGGTCGTGCCAATTGTCAACCCCGCACGTTCATAGCCATAAAATCGTGCTGACC ATGAGCTGCGATTACCTCCGGGCATTATTTCGATCCGGGATGCATGAGAGCTTCTCCGAC GTTATCAGGGTTCCGCTCGGGTGGCAAGCGCTGGACAAGCTGGCTCACTGGTTCTACTCG GGCGAGCTGCCGAAAGCCGCCGTCGACTGCCGATGGAACAACCTGGGCAGCGACGAGCAG CGCGCCCACCTGAACGCCTACGCGGAGCTGTCGTCCCTGTCCGAGTTCTGGTTCCTGGAG GGGGTGAAGGAGGAGAGCCTCGTCGCGGCGTCGTCCCTCCTGGAGTCGAGCACGAGCGCC GCCGCGGTCGGGTTCGTCGCCTTCGCGGCCAGCCTGGGCCAGTGGGAGATGGTGGACGCC GGCGTCCGGAGCGTGGCGCACCTGTACCCCCGGCTCCGGGACTCCGGCGGGCTGGAGCGG CTCGACGACGAGCTGCTCAACATGCTGCGCACCGAGTACGTCCGCTACTCGCAGCACGGA GGCAGGCAGAGGTGACTGAATGATACGTACGTACATGGGGGACCTGAATGCGCGTCGCCG TGCGCTATGTAGATCCCCCCCCAGGGATTGCTGCCATGCTTTCTTCACAGAGAATAATGT GCAATGTAAGTAGTAGCATTATCGTCTGGGTTGCCAGAGCGGAGAGCTGTTTGTTGCTCC TGCCTGCATGGCGCACATGGTCACGCGCAAAGGTGCAGGTATCGGAGGACCGACCGGCCT GCTGACACGCACACGCCGCGCCGATCCATCGCCACGCGCATCACAACGGACACTTTATAT TTTTTTCCCTTCCTTTTCGCGGATCGTCACAACGGACGCCTGGTCACTTGGCTCTGGTGA TCAACGCTAGTTGCCGAAGTCAGTCATCAACGCTCGGCTCAAAGCAAGCAGCTAAGATAG TCCCAACGGAACAACCCGCGGATCTCTCCTCAATCATCATCACGCGAAACGACGGAAGAT AATTAATTAACGGGGCGGCAACATGGTCCGCCAGTCAACAAACAGGGGGGGAGGCCAGCT CGTACGGGAA
Protein Sequence (1004 aa)
>BART1_0-u56048.003 1004 150831_barley_pseudomolecules XXXAGAGAGEHLLTDRVLSLRARLHDALSLGLARSDGHGAKKWQSTDAGIQSHAIKAVTA FLGCLSTEMLRLPPIKESISDILVLLEGVLQTKNVSVLIQAADVSSKLVSSIGNSIRQYS ILEIVSPLSCQLSAHQLPTAISCARAINCILNSLVTARGSTHAEILETLERTNAVASIVS ALQSYTLDVHPLSYLTEMISLLRTILWTWPSSRYHVWSNGDLMGKLAHYCLATETTVVAK VLRLYAALALCGHGATILLKNEVLMAKICELVGTSHPTSTRIEAFKLCHVLLRSSKGCSQ LMTSRCQPIVEGIIDAMRAKDDKLLVTEGCRTALVVLRYAGNHHQCFWSNAIDKVLYTIL TGRCLSSHQSDQVLCDDELFDMVSKNFMDIHPYVWDILGYLVVHCTDKHHPVRKGKGHSL HALISCACLLATDVMRKSSPMKLSKDVQEPALRAVLMMLLSPNGYIQYEASFKLSEGLPY LGDGYLNVLLSSLESNATRSVATSFDSFKIMTNLMNLACLVLSRPYHNLLNKRNPIDVLS TIIKESLHNHIHITRSKVTSHLHFCFDGSSCCCYLGEEWEGENIVLFYGLVVLFNLMRST TLVCVQCKRKLDVGNLCHDCRDQYTEDFLIVLQHALSQSLSSGPKLYIAHILSLFGLCGS PSKLGGKMTSALDNNDLADLELLLSDGESLNAHTAIISARCPKLLPSVKYLLGSDEKVKD EWGKSVYRVQMSDRVDSRALKKILEYTYTGFLMVGDDVVKPVRTLAKYCHLKPLAEMLQK EQPRWNSDYARYDLSAALGPVEHSFSDIILEARSNEEMNCHHGSCQLSTPHVHSHKIVLT MSCDYLRALFRSGMHESFSDVIRVPLGWQALDKLAHWFYSGELPKAAVDCRWNNLGSDEQ RAHLNAYAELSSLSEFWFLEGVKEESLVAASSLLESSTSAAAVGFVAFAASLGQWEMVDA GVRSVAHLYPRLRDSGGLERLDDELLNMLRTEYVRYSQHGGRQR
