- Transcript BART1_0-u56048.002
- Transcript IDBART1_0-u56048.002
- Gene IDBART1_0-u56048
Exon Structure of BART1_0-u56048.002
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 620784789 | 620784689 | + |
| chr7H | 2 | 620785405 | 620785279 | + |
| chr7H | 3 | 620786395 | 620785866 | + |
| chr7H | 4 | 620786624 | 620786494 | + |
| chr7H | 5 | 620787133 | 620786727 | + |
| chr7H | 6 | 620788394 | 620787261 | + |
| chr7H | 7 | 620788805 | 620788653 | + |
| chr7H | 8 | 620789788 | 620788920 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials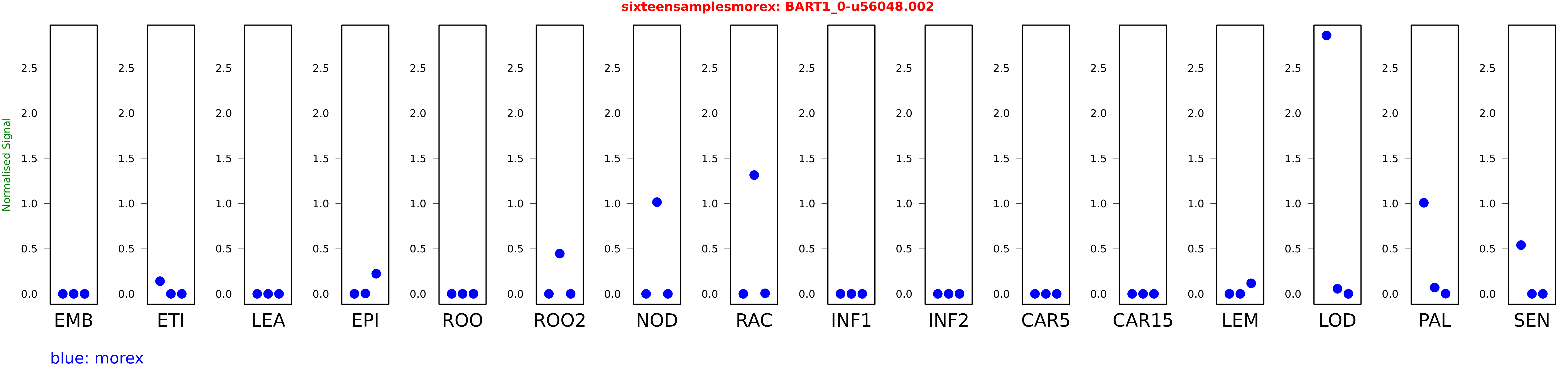
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.0000599324 | 0 | 0 |
| morex | ETI | 0.140425 | 0.000000017474 | 0.000180938 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0.00447771 | 0.222624 |
| morex | ROO | 0 | 0 | 0 |
| morex | ROO2 | 0 | 0.445656 | 0 |
| morex | NOD | 0 | 1.01532 | 0 |
| morex | RAC | 0 | 1.31469 | 0.00631008 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0.00000356781 | 0.00000000138711 | 0.116744 |
| morex | LOD | 2.86037 | 0.0561079 | 0.000000873032 |
| morex | PAL | 1.00816 | 0.0698663 | 0.00149959 |
| morex | SEN | 0.539727 | 0.000000302851 | 0.0000000708582 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os06g46240.1 | +2 | 0.0 | 346 | 173/276 (63%) | protein|BTB/POZ domain containing protein, putative, expressed |
| TAIR PP10 | AT1G04390 @ ARAPORT AT1G04390.1 @ TAIR |
+2 | 5e-69 | 249 | 145/362 (40%) | Symbols: | BTB/POZ domain-containing protein | chr1:1179678-1183615 REVERSE LENGTH=849 |
| BRACH PP3 | Bradi1g32690.2.p | +2 | 0.0 | 1136 | 593/755 (79%) | pacid=32804374 transcript=Bradi1g32690.2 locus=Bradi1g32690 ID=Bradi1g32690.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3452 bp)
>BART1_0-u56048.002 3452 150831_barley_pseudomolecules NNNNNNNGCGCCGGCGCCGGCGCCGGGGAGCATCTCCTCACCGACCGCGTCCTCTCCCTC CGCGCACGCCTGCACGACGCCCTCTCCCTCGGCCTCGCCAGGTCTGATGGCCACGGTGCA AAAAAATGGCAGTCTACTGATGCTGGAATTCAGTCTCACGCGATCAAAGCAGTGACTGCT TTTCTCGGTTGTTTGTCAACTGAAATGCTGCGGCTTCCTCCTATAAAGGAGTCAATTTCG GATATTCTCGTACTCTTGGAAGGTGTTCTTCAGACAAAGAACGTTTCAGTTCTGATCCAA GCAGCAGATGTGAGCTCAAAGTTAGTTTCCAGCATAGGCAATTCGATTCGGCAATACTCA ATTTTGGAGATTGTTTCGCCCCTCTCATGCCAGTTGTCGGCACACCAGTTACCTACGGCT ATCTCATGTGCAAGGGCAATAAACTGCATATTGAATAGCCTAGTGACAGCGAGAGGTTCA ACTCATGCAGAAATTTTGGAAACCTTGGAGAGAACCAATGCAGTTGCTAGTATTGTTTCA GCTCTGCAGAGTTACACTCTTGATGTCCATCCACTGAGTTATCTGACCGAGATGATATCA CTGCTAAGAACCATACTGTGGACTTGGCCTTCCTCAAGATACCATGTATGGAGTAACGGC GACCTGATGGGGAAGTTAGCACATTACTGTCTTGCCACTGAAACAACGGTTGTAGCTAAA GTTCTGAGGCTGTATGCTGCTTTAGGTACATCTGTTTGCTTTATGTGGCCATGGAGCTAC GATCCTTCTCAAGAATGAAGTCTTAATGGCTAAGATTTGTGAGCTTGTGGGGACGTCCCA TCCGACTAGTACTAGGATTGAAGCGTTCAAACTCTGCCATGTTCTTTTGAGATCTTCGAA GGGCTGTAGTCAGTTAATGACTTCACGTTGCCAGCCTATTGTTGAAGGCATAATCGATGC AATGCGTGCAAAGGATGATAAATTGTTAGTAACAGAGGGGTGTCGTACTGCACTGGTGGT CCTCCGTTATGCTGGGAATCATCATCAATGCTTTTGGTCTAATGCCATTGATAAAGTATT ATATACCATTCTTACGGGCCGCTGCCTCTCCTCGCATCAAAGTGATCAAGTTTTGTGCGA TGATGAGCTATTTGATATGGTTTCCAAAAACTTCATGGATATACATCCATACGTCTGGGA TATACTTGGATATCTAGTGGTGCATTGCACTGACAAGCACCATCCTGTAAGGAAAGGGAA AGGTCATAGCTTGCACGCACTGATATCTTGTGCTTGCTTGCTGGCCACAGATGTTATGCG CAAAAGCAGCCCCATGAAATTGTCCAAAGATGTGCAGGAGCCGGCTTTGAGAGCAGTTCT GATGATGCTTCTCTCTCCCAATGGATATATTCAGTATGAGGCAAGTTTTAAACTTTCAGA AGGTCTTCCGTATTTGGGTGATGGCTATTTGAACGTTCTATTGTCATCATTAGAATCAAA TGCTACGAGAAGTGTTGCCACATCTTTTGACAGTTTCAAAATTATGACCAACCTCATGAA CCTAGCATGTTTGGTGCTGTCACGACCTTACCATAATCTCCTGAACAAGAGGAATCCTAT AGATGTGTTGTCCACCATTATCAAGGAATCTCTCCACAATCATATACATATTACGAGGTC AAAAGTCACTTCCCATCTGCATTTTTGTTTTGATGGAAGTTCATGCTGTTGCTATCTCGG TGAAGAGTGGGAGGGTGAAAACATAGTTTTATTCTACGGTCTTGTTGTGCTGTTTAATCT TATGAGGAGCACTACTTTAGTTTGTGTTCAGTGTAAGAGGAAGTTGGATGTTGGAAATCT GTGCCATGACTGCAGAGATCAATATACAGAAGATTTTCTTATAGTTCTTCAACATGCTCT AAGTCAAAGCTTGAGTTCTGGACCGAAGCTCTATATTGCACACATCCTGAGTTTGTTTGG CCTCTGTGGTTCTCCAAGCAAGTTGGGCGGGAAGATGACTAGTGCATTAGATAATAACGA TCTAGCTGATCTGGAACTTTTGCTTTCAGATGGTGAATCTCTAAATGCTCACACAGCCAT CATATCAGCGAGGTGTCCTAAGCTGTTGCCATCTGTAAAATATCTACTTGGCAGCGATGA AAAAGTTAAAGATGAATGGGGAAAATCAGTGTACCGAGTTCAGATGTCTGATCGTGTAGA TAGTCGCGCCTTGAAGAAGATTTTGGAGTACACATATACTGGTTTTCTGATGGTAGGCGA TGACGTTGTTAAACCAGTAAGGACACTTGCGAAGTATTGTCACCTGAAACCATTGGCAGA AATGCTTCAGAAAGAGCAACCCAGATGGAACTCCGATTATGCCAGATATGATCTTAGTGC TGCACTTGGACCTGTTGAGCATTCATTCTCGGATATAATTTTGGAGGCTAGGTCAAATGA GGAAATGAATTGTCACCATGGGTCGTGCCAATTGTCAACCCCGCACGTTCATAGCCATAA AATCGTGCTGACCATGAGCTGCGATTACCTCCGGGCATTATTTCGATCCGGGATGCATGA GAGCTTCTCCGACGTTATCAGGGTTCCGCTCGGGTGGCAAGCGCTGGACAAGCTGGCTCA CTGGTTCTACTCGGGCGAGCTGCCGAAAGCCGCCGTCGACTGCCGATGGAACAACCTGGG CAGCGACGAGCAGCGCGCCCACCTGAACGCCTACGCGGAGCTGTCGTCCCTGTCCGAGTT CTGGTTCCTGGAGGGGGTGAAGGAGGAGAGCCTCGTCGCGGCGTCGTCCCTCCTGGAGTC GAGCACGAGCGCCGCCGCGGTCGGGTTCGTCGCCTTCGCGGCCAGCCTGGGCCAGTGGGA GATGGTGGACGCCGGCGTCCGGAGCGTGGCGCACCTGTACCCCCGGCTCCGGGACTCCGG CGGGCTGGAGCGGCTCGACGACGAGCTGCTCAACATGCTGCGCACCGAGTACGTCCGCTA CTCGCAGCACGGAGGCAGGCAGAGGTGACTGAATGATACGTACGTACATGGGGGACCTGA ATGCGCGTCGCCGTGCGCTATGTAGATCCCCCCCCAGGGATTGCTGCCATGCTTTCTTCA CAGAGAATAATGTGCAATGTAAGTAGTAGCATTATCGTCTGGGTTGCCAGAGCGGAGAGC TGTTTGTTGCTCCTGCCTGCATGGCGCACATGGTCACGCGCAAAGGTGCAGGTATCGGAG GACCGACCGGCCTGCTGACACGCACACGCCGCGCCGATCCATCGCCACGCGCATCACAAC GGACACTTTATATTTTTTTCCCTTCCTTTTCGCGGATCGTCACAACGGACGCCTGGTCAC TTGGCTCTGGTGATCAACGCTAGTTGCCGAAGTCAGTCATCAACGCTCGGCTCAAAGCAA GCAGCTAAGATAGTCCCAACGGAACAACCCGC
Protein Sequence (740 aa)
>BART1_0-u56048.002 740 150831_barley_pseudomolecules MAKICELVGTSHPTSTRIEAFKLCHVLLRSSKGCSQLMTSRCQPIVEGIIDAMRAKDDKL LVTEGCRTALVVLRYAGNHHQCFWSNAIDKVLYTILTGRCLSSHQSDQVLCDDELFDMVS KNFMDIHPYVWDILGYLVVHCTDKHHPVRKGKGHSLHALISCACLLATDVMRKSSPMKLS KDVQEPALRAVLMMLLSPNGYIQYEASFKLSEGLPYLGDGYLNVLLSSLESNATRSVATS FDSFKIMTNLMNLACLVLSRPYHNLLNKRNPIDVLSTIIKESLHNHIHITRSKVTSHLHF CFDGSSCCCYLGEEWEGENIVLFYGLVVLFNLMRSTTLVCVQCKRKLDVGNLCHDCRDQY TEDFLIVLQHALSQSLSSGPKLYIAHILSLFGLCGSPSKLGGKMTSALDNNDLADLELLL SDGESLNAHTAIISARCPKLLPSVKYLLGSDEKVKDEWGKSVYRVQMSDRVDSRALKKIL EYTYTGFLMVGDDVVKPVRTLAKYCHLKPLAEMLQKEQPRWNSDYARYDLSAALGPVEHS FSDIILEARSNEEMNCHHGSCQLSTPHVHSHKIVLTMSCDYLRALFRSGMHESFSDVIRV PLGWQALDKLAHWFYSGELPKAAVDCRWNNLGSDEQRAHLNAYAELSSLSEFWFLEGVKE ESLVAASSLLESSTSAAAVGFVAFAASLGQWEMVDAGVRSVAHLYPRLRDSGGLERLDDE LLNMLRTEYVRYSQHGGRQR
