- Transcript BART1_0-u53703.103
- Transcript IDBART1_0-u53703.103
- Gene IDBART1_0-u53703
Exon Structure of BART1_0-u53703.103
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 385862497 | 385861743 | - |
| chr7H | 2 | 385862822 | 385862587 | - |
| chr7H | 3 | 385863972 | 385862887 | - |
| chr7H | 4 | 385864284 | 385864078 | - |
| chr7H | 5 | 385865991 | 385865930 | - |
| chr7H | 6 | 385866338 | 385866227 | - |
| chr7H | 7 | 385866611 | 385866409 | - |
| chr7H | 8 | 385867084 | 385867000 | - |
| chr7H | 9 | 385867226 | 385867200 | - |
| chr7H | 10 | 385867533 | 385867389 | - |
| chr7H | 11 | 385872217 | 385871179 | - |
| chr7H | 12 | 385872364 | 385872313 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials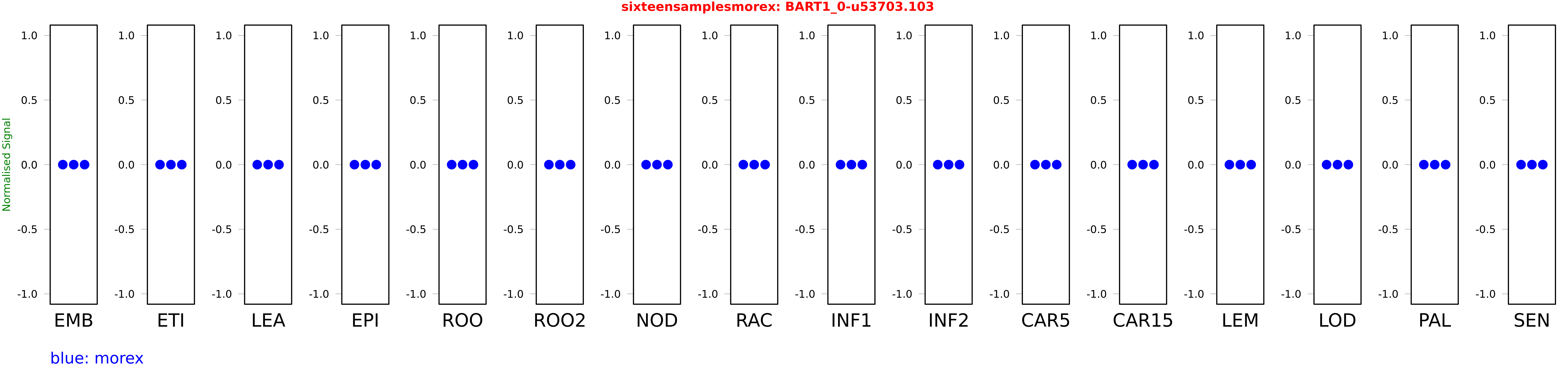
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0 | 0 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0 | 0 | 0 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os08g06110.2 | +1 | 0.0 | 545 | 352/496 (71%) | protein|MYB family transcription factor, putative, expressed |
| TAIR PP10 | AT2G46830 @ ARAPORT AT2G46830.1 @ TAIR |
+1 | 6e-38 | 152 | 71/91 (78%) | Symbols: CCA1 | circadian clock associated 1 | chr2:19246005-19248717 FORWARD LENGTH=608 |
| BRACH PP3 | Bradi3g16515.1.p | +1 | 0.0 | 591 | 375/491 (76%) | pacid=32817815 transcript=Bradi3g16515.1 locus=Bradi3g16515 ID=Bradi3g16515.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (4009 bp)
>BART1_0-u53703.103 4009 150831_barley_pseudomolecules CCGGTGGCGGCAGAGAGAGGAGCTCCTGCGCCCGGCGCGGCGGTCGGTTGAGGCCGCCGC GCGGAGGGCGGGCTGGGCGGGCGACGTGGCCACCGGGGTCAGCTTGCTTGCTGCTCGCCC GAGGTAACCTAGAACTAAGAACCATACAAGTTTCCAGAAATTCGTGCTTCCACTGGACGA CGCGATTGATATATACGCTCCCCTCGCAAGTCCGTTCCTAGGTGTGCTAGTTTGTCGACT GAAAACTGTGCAGCAAATGAGCCCACGCTTTTTTTTTTCTGTGCCTGGTCGGTCTCTGTG GTTGCAGTGTTTGATTCCCGTTGCTCTGCTCCAATGATCATGGTGCACAAGCTCAGAATT CTGAGCGCTTCAACTGCATCCTTTTTCTTGCTCTAGTTAGTCCTGTCCATTTTACTGTGG CTAGGAATAAAATTAATAGTGATTCTATGGTCCAACTTCTTAGAAAACTGTATGGTCGAC TAGTAAACTTAGGTACGTGTTTTTTTATCTTGCTTTTTTAGCTTTCACGGGTTTGAGGTG TGCTTTTATCAGATCTCTGATCTAGTATGGTGTTGTTTGTGCTCTGAAGTTGTTTAAAAA GAACGTGTGTAAGTTAGGCTTCTGAGTTTGCTATGCAGGGAGAGAACAGCTTATTTTTTT TATAAGCAGGTGAGTTAAGTTAATTTTGGATTTGCTATGCTGAAAAGAGAGCAGCTTGAT CTAGTTTAACTAAGCAGGTGAAGTAATTTGACTAAAATAGATGCTATAGTGAATCGTGCT AAGAAGAAAATATATTTTTTATCTGCTATCAAACATTCATGTTTTTTTTTGGTTGTTGTT ATGTCAATATATTTGCTATGATAAAATATCGGGCTGGCTTAAATTCTACTGTTTGTTTCG GAAAGTTCAGCAATCCTAAAGCAACATATGCATATTCAGGTATGTTGCCAAAAAAATATA TGTTTCTATTAGTATTAGTTACTCGTTTTGCTTCCTAGTAAGCTCATAGATATGTTTGCA GGCATACTATGTTGTCCTTAAATAACGTGGTTCAGGTTGTGGGAGATAAACTCATGGATG CTCCTTCAAAGGTGTAGGAAGTGTTAAGGGGATACATAATCGGTTTTGAGCTGCGAGGCC CTGCATTTCTTCTGCCTGCTGTTTGATTCACGGCCCACTCTTTTCCTTCTATCCATTCAA GCAAAGTACTGGAATTTCTGAGCCCAATTTATTCAGGTCATGGGATTTGTTTGTCCTTAG CAGAGTTGAAGATAAAGAGAGGAAGGCTAAACTCTGCTGATCCGGTTCATGCGAGCAATG AGCCAACTGCAGTAGAGGTTTACAGTAGAGAGGTTTAGGGGGAGGTATGAGATACTACTC GCGTCAGGTCCAGCGTTTGGCTGGAGACACAGAAGGAGGAGTGCTGTTGTTTGTAGACGC CCTCAACTCCAAGTAACAAAGCGACCAGTTGTCAGCACTTCCTTTTGTTTTCGCATTTCC CTGGAATTGGAGATGGAGATAAATTCTTCGGGTGAGGAAACGGTGATAAAGGTGCGAAAG CCGTACACAATAACAAAACAGCGGGAGCGGTGGACTGAGGCAGAGCACAAACGGTTCCTT GAAGCCCTCAAACTGTATGGCAGAGCTTGGCAGCGCATAGAAGAGCATGTTGGGACAAAG ACGGCCGTGCAAATCAGAAGCCATGCTCAGAAGTTCTTCACAAAGTTGGAAAAGGAAGCT ATCAACAATGGTACTTCTCCAGGACAAGCTCATGATATAGACATACCTCCACCACGGCCT AAAAGAAAACCTAACTGTCCATATCCTCGAAAAGGTTGTCTCAGCTCTGAGACACCCACC AGAGAAGTTCCAAAATCAAGTGTTAGCTTGAGCAATAGCAATTCACAAATGGAAAGCAAT GGAACTCTTCAGGTCACCAGCACTCAGAAACTTCAAAGGAAGGAGTTGTCTGGAAACGGC AGTTGCTCAGAAGTTATTAATATCTTTAGAGAAGCACCATCTGCCTCATTTTCTTCCTCT AACAAGAGCTCTTCAAATCATGGTGTCTCTGGGGGAATTGAACCGACTAAAACAGAAATC AAAGATATGGCAGCCATGGAAAGGAAATCTACTTCCGTTGATGTGGCGAAGGATGTAAAA GATATTAATGACCAGGAAATGGAAAGGAACAACAGAGTCCACATCAGTTCTAAATATGAC CGTTCTCATGAAGATTGTTTGGATAGCTCAATGAAACACATGCAGTTGAAGCCAAATACT GTGGAGACAACATACACGGGTCAACATGTTGCAAGTGCTCCACTCTACCAAATGAATAAG ACTGGGGCAACTGGCACTCCAGACCCTGGAACTGAAGGAAGTCATCCTGATCAAACGAAT GATCAAGTGGGAGGAGCTAATGGAAGTATGGACTGCATCCATCCAACACTTCCCGTGGAT CTAAAATTTGGCAGCAGCTCCACAGCGCAGCCCTTTCCCCACAACTATTCAGGCTTTGCA CCAACGATGCAATGCCAGTGCAACCAAGATGCCTACAGGTCATCTGTTGATATGTCGTCC ACCTTCTCCAACATGCTCGTTTCCACATTGTTATCAAACCCCACAGTACATGCAGCTGCA AGGCTTGCAGCATCATACTGGCCAGCAGCAGACAGCAACATCCCTGTCGATCCAAATCAA GGAATTTTTGCTCAGAATGCTCAAGGAAGACATATTGTTTCTCCTCCAAGCATGGCTTCT GTGGTAGCAGCTACAGTTGCTGCGGCTTCGGCATGGTGGGCAACACAAGGTCTTCTCCCT CTTTTTGCTCCCCCCATGGCTTTTCCATTTGTCCCAGTTCCTACCGCTTCCTTTCCCACA GCGGATGTCCAGCGAGCTACAGAGAACTGCCCAGTGGACAACGCACCAAAGGAATGCCAA GTAGCTCAGGGGCAAGGTCAACCTGAAGCTATGATAGTTGTAGCATCTTCTGGGTCCGGC GAGAGTGGAAAAGGAGAGGAGCTGAAACAAGTGATGCTTTGGGCAACAAGAAGAAGCAGG ATCGCTCTTCATGTGGTTCCAACACACCATCAAGTAGTGATGTAGAGGCAGAACATGTTC CTGAGAACCAAGATCAAGCTAACGACAAGACACAGCAAGCATGTTGCAGTAATTCTTCAG CTGGTGACATGAACCATCGCAGGTTTAGGAACATTTCAAGCACGAATGATTCATGGAAGG AAGTTTCCGAGGAGGGTCGTATGGCTTTCGATAAACTGTTCAGTAGAGGAAAGCTTCCCC AAAGCTTTTCTCCTCCACAAGCAGAAGGATTGAAGGTGGTTCCCAGAGGGGAGCAAGATG AAGCTACTACGGTGACGGTCGACCTCAACAAGAGTGCTGCAGTTATGGACCATGAACTTG ACACATTGGTTGGGCCAAGAGCTTCCTTTCCCATTGAATTGTCACACCTGAATATGAAAT CCCGCCGGACAGGCTTCAAACCTTACAAGAGGTGCTCGGTGGAAGCAAAGGAGAATAGGG TGCCGGCTGCTGACGAGGTTGGTACCAAAAGGATTCGCCTTGACAGCGAACCCTCCACGT GATTTACTTCCCACGACATGATTACCAGCCTGCACAAGTAGTGTATTTTCAAGAAATTGC GGTATTTACATCTAAGCTACTATAGGACTTGCCAGTCCTTGCAATGCAGTGTTTCACAGA ACTCAATTTGTTATGTGTTGTATGTTAATGCTTGCCGAGCAGCCCTTTTAGACTGTCAAT TAAGCATTAAGCACCTATGCATGCTACTTTATTGGTAAAAAAGTAATTCTGCTTATTTTT CTGTGATGAAGGATAATACTTGTAATTTTGCTTCATTTTACTGGATCATGTAGATTTAAT TCCCGAAGGGAGAGCATGATCGCCTTGTGTTATTTTTATTTTCTTGAGAAAAATATTGTG CTATGTATATGCACCCTAGATTTGAAGTTTCTTGGATGGTCCAGCTTTC
Protein Sequence (530 aa)
>BART1_0-u53703.103 530 150831_barley_pseudomolecules MEINSSGEETVIKVRKPYTITKQRERWTEAEHKRFLEALKLYGRAWQRIEEHVGTKTAVQ IRSHAQKFFTKLEKEAINNGTSPGQAHDIDIPPPRPKRKPNCPYPRKGCLSSETPTREVP KSSVSLSNSNSQMESNGTLQVTSTQKLQRKELSGNGSCSEVINIFREAPSASFSSSNKSS SNHGVSGGIEPTKTEIKDMAAMERKSTSVDVAKDVKDINDQEMERNNRVHISSKYDRSHE DCLDSSMKHMQLKPNTVETTYTGQHVASAPLYQMNKTGATGTPDPGTEGSHPDQTNDQVG GANGSMDCIHPTLPVDLKFGSSSTAQPFPHNYSGFAPTMQCQCNQDAYRSSVDMSSTFSN MLVSTLLSNPTVHAAARLAASYWPAADSNIPVDPNQGIFAQNAQGRHIVSPPSMASVVAA TVAAASAWWATQGLLPLFAPPMAFPFVPVPTASFPTADVQRATENCPVDNAPKECQVAQG QGQPEAMIVVASSGSGESGKGEELKQVMLWATRRSRIALHVVPTHHQVVM
