- Transcript BART1_0-u53291.018
- Transcript IDBART1_0-u53291.018
- Gene IDBART1_0-u53291
Exon Structure of BART1_0-u53291.018
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 367196746 | 367196666 | + |
| chr7H | 2 | 367196913 | 367196832 | + |
| chr7H | 3 | 367197114 | 367197079 | + |
| chr7H | 4 | 367197307 | 367197240 | + |
| chr7H | 5 | 367197953 | 367197465 | + |
| chr7H | 6 | 367198300 | 367198088 | + |
| chr7H | 7 | 367199069 | 367198377 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials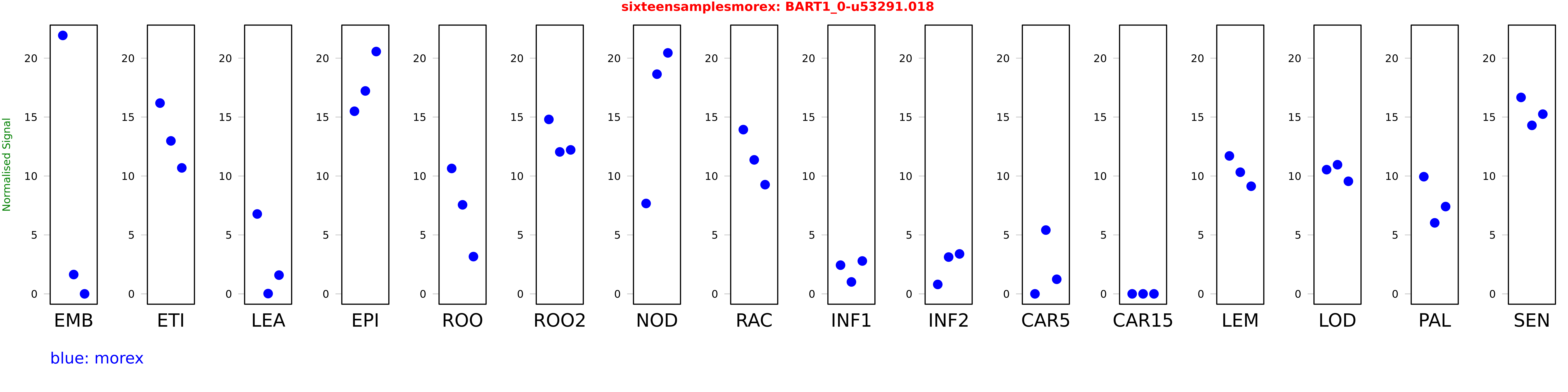
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 21.9282 | 1.63842 | 0 |
| morex | ETI | 16.1858 | 12.9781 | 10.6879 |
| morex | LEA | 6.77723 | 0.0217707 | 1.58894 |
| morex | EPI | 15.4944 | 17.2173 | 20.5602 |
| morex | ROO | 10.6408 | 7.55417 | 3.16092 |
| morex | ROO2 | 14.8022 | 12.0436 | 12.2157 |
| morex | NOD | 7.66923 | 18.6407 | 20.445 |
| morex | RAC | 13.9351 | 11.3692 | 9.26495 |
| morex | INF1 | 2.43505 | 1.01177 | 2.78902 |
| morex | INF2 | 0.799989 | 3.12171 | 3.38824 |
| morex | CAR5 | 0.000152614 | 5.41179 | 1.23764 |
| morex | CAR15 | 0 | 0 | 0.00000122523 |
| morex | LEM | 11.7038 | 10.3216 | 9.13172 |
| morex | LOD | 10.5388 | 10.9562 | 9.55291 |
| morex | PAL | 9.94057 | 6.0241 | 7.40631 |
| morex | SEN | 16.6683 | 14.298 | 15.2487 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os08g12760.2 | +1 | 0.0 | 649 | 319/492 (65%) | protein|YT521-B, putative, expressed |
| TAIR PP10 | AT3G13060 @ ARAPORT AT3G13060.2 @ TAIR |
+1 | 6e-103 | 323 | 180/378 (48%) | Symbols: ECT5 | evolutionarily conserved C-terminal region 5 | chr3:4180625-4183632 FORWARD LENGTH=634 |
| BRACH PP3 | Bradi3g18190.1.p | +1 | 0.0 | 734 | 361/496 (73%) | pacid=32821513 transcript=Bradi3g18190.1 locus=Bradi3g18190 ID=Bradi3g18190.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (1662 bp)
>BART1_0-u53291.018 1662 150831_barley_pseudomolecules GTAGGATATATCAACCCTTCAGGGCAATGGGAAGAGTATCCCTATTATGTGAGTATGGAA GGAGTCCATTCTGCGTCACCTTCAATCATGCTATCTCCTGGGTATGCGAACAACCCCCAA ATGATGTATGGAGCTTATTCTCCTGTTTCAACTGTTGGAGAGGCAGTGTACCGCCAACCA TTTGGGTCATACAACAGAGCGGCAACACAACCAAGCAATGCTTCGGGACTATTTGGGCAA GGGAATGCACCATTGGCTTTTGGAATGCAGCATGGAACAATGTATAATTCTGGATCCTAC AAGGCACACCAACAAGCTAGTAAATATGGAGGCACTACTCCTAATTGGGGTTCTGCCGGT CGTAGATTTAATAATTTTGACTGCAACTCCAGTCAACAAAGGGGGAGCATGCCATTCAAT ATCCAGAATGGTGCTCTGGAGTTTCTGAACGAGCAAAACCGCGGCCCACGTGCTGCAAAA CCAAAGAAACAAGACAGAGACAATTCTTTAGTAGATGACAAAAGCGAGAAAACCACTCTG CTTATTGACAGTGAACTGTACAATCGTCCTGATTTTGCCACGGAGTACAAGGATGCGATT TTTTTTGTAATTAAATCATACACAGAGGACCATGTACATAGGAGCATTAAGTACAATGTC TGGGCCAGCACAGCTAGTGGGAATAGAAAGCTGGATTCAGCTTATCGTGCGGCCAAACAG AAAGAAGACCATTCCCGTGTTTTCTTGTTCTTCTCGGTTAATGGCAGTGGTCAGTTCTGC GGTGTTGCTGAGATGATTGGACCTGTTGACTTCGACAGGAGCGTTGATTATTGGCAGCAA GACAAGTGGAGTGGCCAGTTTCCTGTGAAGTGGCACATTATTAAGGATGTCCCAAACACC CTACTCCGGCACATCACTCTTGAGAACAATGACAACAAACCTGTAACAAATAGCAGGGAC ACTCAGGAGGTCAAGCTAGAGTATGGTCTCCAGATGCTAAGCATATTCAAGAATCATGAG GCCGAGACAACCATTGTGGAGGACTTCGACTTCTATGAGCAACGGGAGGAAGCCTTGAAG GAAAATAGACGTCAGCAGCAGCCTGGCAGCGCAGAGTCCCTGAAGCCAACAGATGCTAAG GCTGTAGGAAGTTCTGTTACTCATATCGTGGATTCATTTTCTCGAAGCGTCCAGCTGAAA GAGATCGAGAAAAGTGATAAGAGAACTGATGGTGCCATCTCAGTGAATGGTTCACAGACA GCTACCGTCAAGGCCGAAAAAAGCACTGCAAACAAAACTACAGGTTCTGTGGAGGAGAGC AGTTGATTACCTCCTGCCATTTACTAGTTTGGAGCAGCAGTTGGATCCATATGATTTGAT GCTCGTCTGGAGTTGAGAAGGGAAAAAGCTGCCGATTCGGGTTTAGATTTGGGCTGCTGC TGCTTGCAGTTGTGAACATCATACTCCTTATTTCGCCTCCATTTAGGTTATAGTTTTTCA ATATCATAGTTTCAAAATTCTTGTTTGTGGATCCTTGCTTCTGTGTTCTTTCTTGTTTTC TTCTCTCTTTGTTTTACTTGCGTTGTGTCGTGAATGCTGTTCAGTTTTAGTGGCCTTCTC ATCACTTTTGGTTTACCGTCTGTCATAAATGCTGCCCAGTTG
Protein Sequence (441 aa)
>BART1_0-u53291.018 441 150831_barley_pseudomolecules VGYINPSGQWEEYPYYVSMEGVHSASPSIMLSPGYANNPQMMYGAYSPVSTVGEAVYRQP FGSYNRAATQPSNASGLFGQGNAPLAFGMQHGTMYNSGSYKAHQQASKYGGTTPNWGSAG RRFNNFDCNSSQQRGSMPFNIQNGALEFLNEQNRGPRAAKPKKQDRDNSLVDDKSEKTTL LIDSELYNRPDFATEYKDAIFFVIKSYTEDHVHRSIKYNVWASTASGNRKLDSAYRAAKQ KEDHSRVFLFFSVNGSGQFCGVAEMIGPVDFDRSVDYWQQDKWSGQFPVKWHIIKDVPNT LLRHITLENNDNKPVTNSRDTQEVKLEYGLQMLSIFKNHEAETTIVEDFDFYEQREEALK ENRRQQQPGSAESLKPTDAKAVGSSVTHIVDSFSRSVQLKEIEKSDKRTDGAISVNGSQT ATVKAEKSTANKTTGSVEESS
