- Transcript BART1_0-u51812.012
- Transcript IDBART1_0-u51812.012
- Gene IDBART1_0-u51812
Exon Structure of BART1_0-u51812.012
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 140679678 | 140679396 | + |
| chr7H | 2 | 140680417 | 140680127 | + |
| chr7H | 3 | 140680803 | 140680682 | + |
| chr7H | 4 | 140681781 | 140681165 | + |
| chr7H | 5 | 140683083 | 140681885 | + |
| chr7H | 6 | 140683501 | 140683422 | + |
| chr7H | 7 | 140684053 | 140683958 | + |
| chr7H | 8 | 140684216 | 140684143 | + |
| chr7H | 9 | 140684701 | 140684566 | + |
| chr7H | 10 | 140685517 | 140684971 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials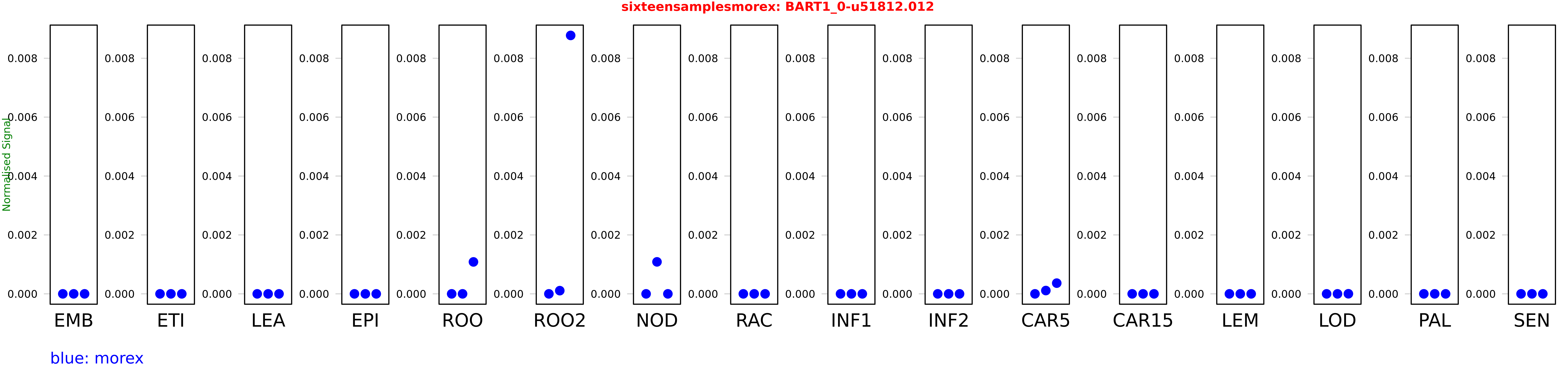
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0.000000159686 | 0.00108438 |
| morex | ROO2 | 0 | 0.000108714 | 0.00877274 |
| morex | NOD | 0 | 0.00108604 | 0 |
| morex | RAC | 0.000000111802 | 0 | 0 |
| morex | INF1 | 0.0000000145695 | 0.00000000737235 | 0.00000000198976 |
| morex | INF2 | 0.0000000420736 | 0.00000000673279 | 0.0000000177629 |
| morex | CAR5 | 0 | 0.000112483 | 0.00036387 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0.00000000019677 | 0.000000124986 | 0.00000307209 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0 | 0.000000695879 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os06g19470.2 | +1 | 0.0 | 798 | 540/826 (65%) | protein|WW domain containing protein, expressed |
| TAIR PP10 | AT3G13225 @ ARAPORT AT3G13225.1 @ TAIR |
+1 | 1e-34 | 142 | 105/183 (57%) | Symbols: | WW domain-containing protein | chr3:4258928-4261728 REVERSE LENGTH=771 |
| BRACH PP3 | Bradi1g43210.1.p | +1 | 0.0 | 934 | 618/825 (75%) | pacid=32799652 transcript=Bradi1g43210.1 locus=Bradi1g43210 ID=Bradi1g43210.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3445 bp)
>BART1_0-u51812.012 3445 150831_barley_pseudomolecules CTCAACCGAAGCGCCCACCCCGGACACCCGCCTACCGCCAGCTCCCTCCACTTCGCCTCT CCTCCTGATTCGCACCGCTTCCCTTTTTGCCGTAAAACAAAACACCCCGACGTCACCCTC CAAACCCTAACCCTAGAATCTAGAAGACGTCCATCTCCACCCCCGGCGCGCGCCCGACTA CTCGGGGCAGCGGCGATGGGGAGGCGGAAGGAGCGCCGCCTGGCGGCCAAGGCGGCCTCG GGCCGTAGGGTCAAGCTCGACCTCTTCCTCGATCCTTCCCCCGGGGAGGCATCTACTAAA GAGGGAGTAGGGGAGGAAAACCGCGAGCAGCAAACTGGTGTTCCCACTTCACCATCTTCG TCAGGTGGGTTCAATCAATTTCCTTCCAAGGGATATGTAAAGACGGCACATGCCTATGGA ATGTAAATTTGGTGGAAATTTGGAATTTTGTTCAAGCATGTGTATGCTCCAAATTTCTGG TTTGAGGCACATGTAGGGTTCCCGGCCTGCAAGGAACACTGAGTAAATTCTTCAGATCAG GCTGCCACTGTCAAAACTGGGTGTGTTCCTACCGATAAGAAGGAAAATCCTCTAGCGTTG CTTGGGCAATATAGTGATGATGAAGAAGAGGATGAAGTAGCAACAGATCAACCCAATGGT GAAACTAAGGGAAGTTCAACTGATGCAAGTGCCAAGGTTTTTAACGATCATGGTGATGTA ACTGGGGATAAAGGCGATGCAGATAGTGAACTGCCTGCTTCTGTTAGTGTTCAGCAGGAA GCATCTCAAGCCGATGATGTAAAAACTATCACAGAAAATGTTGCTGAGGAATTTACTGTT GCCCCTGAGCCAACTCTGGAAAATGAGTGTGTGACAGTAATGGAAGCTGTCCCAGATTCA TCTGGCATGCAAATCGTGGGTGACATTGGTGGGAATTGGAAGGCTATAATGCATGAACAA AGTAATCAGTGTTACTACTGGAACACAGTTACAGGAGAAACTTCTTGGGAGATTCCTGAT GGATTAGCTTCGGGGGTTGCTGGTGATGGAGTCACTTCTGCATCTGTGCCTACTCATATG GGCTACTCTGTAGAAGCTCAAGCACATGTCCTTCCCCACAGCAATGTTGAAGCATATCCT AGTGATGTATCTGTTGGGAATGGCACAGCAACCTATACTGCTATGGGAACTTATACTCAT GATGCTTATGCTTACACTGGAGCCGTCGCTAGTCATGAGTCAGTGGACATTGACCCCTTG CAGCTTGCAAAATATGGTGAGGATTTACTGCAAAGATTGAAGCTGCTGGAAAGGCCACAT GTTGCCATTGACAGTCTTGAGTTGATAAAAAGAGAAATTGAGATACGAGTAGCAGACTGT AATGCACTCTCCTCGTATGGATCTTCTTTACTTCCATTGTGGTTGCATGCTGAGGTGCAC CTTAAGCAACTAGAACTTTCAGTTTCCAAGTTTGAAGCTAGCTACACTACCAGACATGGA TATCTGGAGACGGTGGATGCAGGACACAAAGCACCTAATGAAGCTAAAGTTATGGCACCC TCCGAGGGTGAGCATTTGAAAGTTGATGTTAGCACTCTGGGAATTGGTACTGGCGATGAA AATATTAAGGTTGAGGAACCATGTACAACATCATCTGCTCAGAACTCACAATGTGTGGCA GCAGCTATTTTAAGAGTTGAATCAGACAGTGATGAAGATATGGACGTGGACATGGAGGTC GATGAAGAAGGTGTTGAAGAGCACGGCGGATTCACTTCTATGCCAAATAAGGAGCATCCT TCATCAGAGCAAGTACGATCACCAGCTTTGTCATCATTGGAAGACTCTGCTCCTCCCCAA CAGGATAATGACATTCCTCCTCCACCACCACCACCAGAAGAGGAATGGATTCCACCTCCA CCACCGGAGAATGAACCAGCTCCTCCACCTCCTCCTGAGCCTGAAGAGCCTGCTGTGTCA TTTGTTCATGCTGATACACTTCCTCAGCCATACGGAGGTCAAGCAAACTTGGGTTATATG CTTCCAGGAATGGAGTACTATCCCGCTTCTGGTACAGATGGCACAAATGCCAGCTACTAT ATGCAAGCGAGCGATTCTCATATTCTTCAATCGCAGCAGCATTCTTACTACGCACCGTTG TCTGCAAGTGGCGTATCTATTCCTGTTGAGACCACATCGATCCCCCCAGTACCAGGTTCT TATTATAGCTATCCTTCTGTCACTATGGCTGCCACTGAAGTAGCGGCTGAATCTTCTGGA TACTATGGTTCATCAACCTCTGCCATTTCTAGCGGTGAATTAGATAACAAAACAAGCTCA GCCTCCCTTGTTTCAAATAGTAATGTGAATCCTGTGGAGTCTGATAAAGTGATATCCAAG GAGCCTACAGTTGTTTCTTTGAGCCAATCAGTAGGGGCAGCCTCAGCTTCAGCACCAACA GTACATGGTAGTTCTACTCTAGCTTCTACTAGTACCACTAATCAGACTAAAGTTCCTCGT ACTAAGAAGCGACCTGTTGCTGTTGCATCATCACTGAGATCTAATAAGAAGGTCTCAAGT CTGGTGGATAAGTGGAAAGCTGCAAAAGAGGAGCTTTGTGATGAAGAGGAAGAGGAGCCT GAAGATGCTTTGGAGTACCTAGAAAGGAAACGTCGGAAGGAAATAGATGGGTGGCGTAAG CAACAAATAGCTAGTGGAGAAGCTAAGGAAAATGCTAATTTTGTTCCCCTTGGCGGTAAC TGGCGTGACCGTGTAAAACGCAAAAGAGCTGAAGCAAAGAAGGAAGCAAAGACTGAATCT GTTTGTGCAGCTGCCGAACAACACAAAGGAGAGCCTGATCTCTCAGAGCTTTCCAAGGGT CTCCCTTCTGGGTGGCAGGCATACATAGACGAGTCTACGAAGCAAGTGTACTATGGGAAC AACCTCACTTCCGAGACTACTTGGGATCGGCCTAGCAAATAAAGATTGAACTAGTTGAAG ATGGTCCAGTTTTGCTCTGTAGATATCCTCATACATCTATACGTTGCCTGTCTGGTTATG AGGCTGTATTGTTTCTTTTATTTTCCACTGGGAGATCGTAGCAGGAAGGATACTTCATTT TGGTATATAGAGGCATCCCTTGTAAGAGTGGCAACTTGAGATCGAATTACTGAAGCAAAA AATACTAGTTAGTTGCTGACCAAGGAAAAATGTGCCTTGTTAAGTGGTGGGAGTCTTTAC CTCGATCTGATACCATTTGCTTTCAAAACAATGTCAATTATATTCGATTTTAGGTTGTGC TAAATTCTAGACTTCTCCAATTATTTAACATTAGAAAGAAAGATGATTTTTAAGTTTTTT TTCACACTTATTTTTTTTTTAAACTTCAAAATGCCCTCGGACATCATTGGCTGGATTTTG TTTACATCATGCGATCTAAGTTTCG
Protein Sequence (700 aa)
>BART1_0-u51812.012 700 150831_barley_pseudomolecules MEAVPDSSGMQIVGDIGGNWKAIMHEQSNQCYYWNTVTGETSWEIPDGLASGVAGDGVTS ASVPTHMGYSVEAQAHVLPHSNVEAYPSDVSVGNGTATYTAMGTYTHDAYAYTGAVASHE SVDIDPLQLAKYGEDLLQRLKLLERPHVAIDSLELIKREIEIRVADCNALSSYGSSLLPL WLHAEVHLKQLELSVSKFEASYTTRHGYLETVDAGHKAPNEAKVMAPSEGEHLKVDVSTL GIGTGDENIKVEEPCTTSSAQNSQCVAAAILRVESDSDEDMDVDMEVDEEGVEEHGGFTS MPNKEHPSSEQVRSPALSSLEDSAPPQQDNDIPPPPPPPEEEWIPPPPPENEPAPPPPPE PEEPAVSFVHADTLPQPYGGQANLGYMLPGMEYYPASGTDGTNASYYMQASDSHILQSQQ HSYYAPLSASGVSIPVETTSIPPVPGSYYSYPSVTMAATEVAAESSGYYGSSTSAISSGE LDNKTSSASLVSNSNVNPVESDKVISKEPTVVSLSQSVGAASASAPTVHGSSTLASTSTT NQTKVPRTKKRPVAVASSLRSNKKVSSLVDKWKAAKEELCDEEEEEPEDALEYLERKRRK EIDGWRKQQIASGEAKENANFVPLGGNWRDRVKRKRAEAKKEAKTESVCAAAEQHKGEPD LSELSKGLPSGWQAYIDESTKQVYYGNNLTSETTWDRPSK
