- Transcript BART1_0-u51812.003
- Transcript IDBART1_0-u51812.003
- Gene IDBART1_0-u51812
Exon Structure of BART1_0-u51812.003
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr7H | 1 | 140679678 | 140679390 | + |
| chr7H | 2 | 140680474 | 140680127 | + |
| chr7H | 3 | 140680803 | 140680682 | + |
| chr7H | 4 | 140681781 | 140681165 | + |
| chr7H | 5 | 140683083 | 140681885 | + |
| chr7H | 6 | 140683501 | 140683422 | + |
| chr7H | 7 | 140684053 | 140683958 | + |
| chr7H | 8 | 140684216 | 140684143 | + |
| chr7H | 9 | 140684701 | 140684566 | + |
| chr7H | 10 | 140685432 | 140684971 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials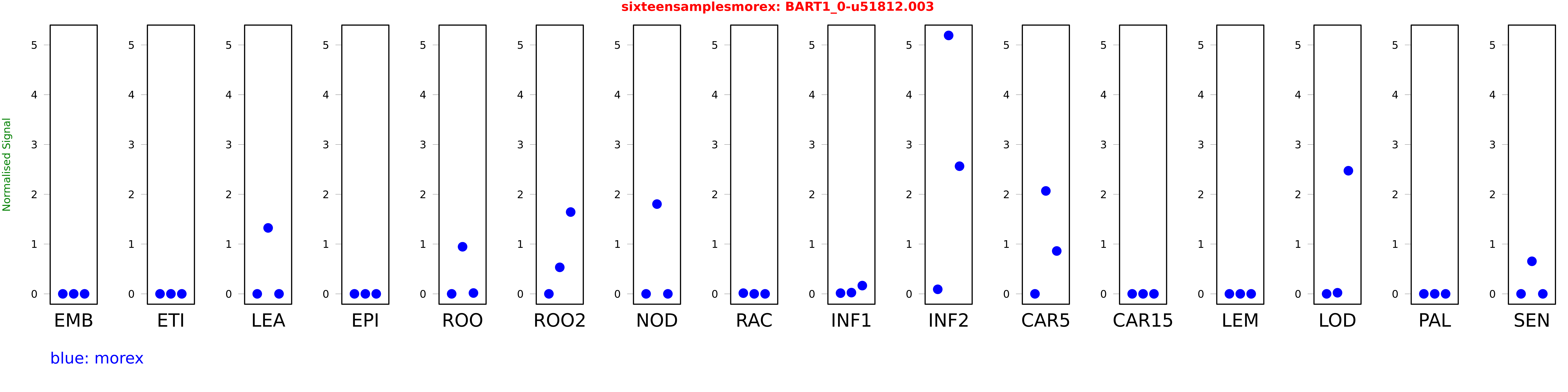
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0.0000000587385 | 0.00000000482541 | 0.0000000925073 |
| morex | LEA | 0 | 1.32438 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0.945917 | 0.0167224 |
| morex | ROO2 | 0.000000713458 | 0.532646 | 1.64308 |
| morex | NOD | 0 | 1.803 | 0 |
| morex | RAC | 0.0143275 | 0 | 0.00000000014673 |
| morex | INF1 | 0.0143594 | 0.0249095 | 0.165785 |
| morex | INF2 | 0.0914722 | 5.19062 | 2.56392 |
| morex | CAR5 | 0 | 2.06718 | 0.860939 |
| morex | CAR15 | 0.000000000681511 | 0 | 0 |
| morex | LEM | 0.00000000402039 | 0.00000000099906 | 0.00000150581 |
| morex | LOD | 0.0000994052 | 0.0221606 | 2.47301 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0.00000472682 | 0.654013 | 0.0000000255294 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os06g19470.2 | +1 | 0.0 | 794 | 536/813 (66%) | protein|WW domain containing protein, expressed |
| TAIR PP10 | AT3G13225 @ ARAPORT AT3G13225.1 @ TAIR |
+1 | 1e-34 | 142 | 105/183 (57%) | Symbols: | WW domain-containing protein | chr3:4258928-4261728 REVERSE LENGTH=771 |
| BRACH PP3 | Bradi1g43210.1.p | +1 | 0.0 | 931 | 615/812 (76%) | pacid=32799652 transcript=Bradi1g43210.1 locus=Bradi1g43210 ID=Bradi1g43210.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3423 bp)
>BART1_0-u51812.003 3423 150831_barley_pseudomolecules GCACAGCTCAACCGAAGCGCCCACCCCGGACACCCGCCTACCGCCAGCTCCCTCCACTTC GCCTCTCCTCCTGATTCGCACCGCTTCCCTTTTTGCCGTAAAACAAAACACCCCGACGTC ACCCTCCAAACCCTAACCCTAGAATCTAGAAGACGTCCATCTCCACCCCCGGCGCGCGCC CGACTACTCGGGGCAGCGGCGATGGGGAGGCGGAAGGAGCGCCGCCTGGCGGCCAAGGCG GCCTCGGGCCGTAGGGTCAAGCTCGACCTCTTCCTCGATCCTTCCCCCGGGGAGGCATCT ACTAAAGAGGGAGTAGGGGAGGAAAACCGCGAGCAGCAAACTGGTGTTCCCACTTCACCA TCTTCGTCAGGTGGGTTCAATCAATTTCCTTCCAAGGGATATGTAAAGACGGCACATGCC TATGGAATGTAAATTTGGTGGAAATTTGGAATTTTGTTCAAGCATGTGTATGCTCCAAAT TTCTGGTTTGAGGCACATGTAGGGTTCCCGGCCTGCAAGGAACACTGAGTAAATTCTTCA GATCAGGCTGCCACTGTCAAAACTGGGTGTGTTCCTACCGGTAATTTCTCTTTTGGCAGC ATCTGAGCGTGGGGATTTCAGGATGATTGTTGCCTCGATAAGAAGGAAAATCCTCTAGCG TTGCTTGGGCAATATAGTGATGATGAAGAAGAGGATGAAGTAGCAACAGATCAACCCAAT GGTGAAACTAAGGGAAGTTCAACTGATGCAAGTGCCAAGGTTTTTAACGATCATGGTGAT GTAACTGGGGATAAAGGCGATGCAGATAGTGAACTGCCTGCTTCTGTTAGTGTTCAGCAG GAAGCATCTCAAGCCGATGATGTAAAAACTATCACAGAAAATGTTGCTGAGGAATTTACT GTTGCCCCTGAGCCAACTCTGGAAAATGAGTGTGTGACAGTAATGGAAGCTGTCCCAGAT TCATCTGGCATGCAAATCGTGGGTGACATTGGTGGGAATTGGAAGGCTATAATGCATGAA CAAAGTAATCAGTGTTACTACTGGAACACAGTTACAGGAGAAACTTCTTGGGAGATTCCT GATGGATTAGCTTCGGGGGTTGCTGGTGATGGAGTCACTTCTGCATCTGTGCCTACTCAT ATGGGCTACTCTGTAGAAGCTCAAGCACATGTCCTTCCCCACAGCAATGTTGAAGCATAT CCTAGTGATGTATCTGTTGGGAATGGCACAGCAACCTATACTGCTATGGGAACTTATACT CATGATGCTTATGCTTACACTGGAGCCGTCGCTAGTCATGAGTCAGTGGACATTGACCCC TTGCAGCTTGCAAAATATGGTGAGGATTTACTGCAAAGATTGAAGCTGCTGGAAAGGCCA CATGTTGCCATTGACAGTCTTGAGTTGATAAAAAGAGAAATTGAGATACGAGTAGCAGAC TGTAATGCACTCTCCTCGTATGGATCTTCTTTACTTCCATTGTGGTTGCATGCTGAGGTG CACCTTAAGCAACTAGAACTTTCAGTTTCCAAGTTTGAAGCTAGCTACACTACCAGACAT GGATATCTGGAGACGGTGGATGCAGGACACAAAGCACCTAATGAAGCTAAAGTTATGGCA CCCTCCGAGGGTGAGCATTTGAAAGTTGATGTTAGCACTCTGGGAATTGGTACTGGCGAT GAAAATATTAAGGTTGAGGAACCATGTACAACATCATCTGCTCAGAACTCACAATGTGTG GCAGCAGCTATTTTAAGAGTTGAATCAGACAGTGATGAAGATATGGACGTGGACATGGAG GTCGATGAAGAAGGTGTTGAAGAGCACGGCGGATTCACTTCTATGCCAAATAAGGAGCAT CCTTCATCAGAGCAAGTACGATCACCAGCTTTGTCATCATTGGAAGACTCTGCTCCTCCC CAACAGGATAATGACATTCCTCCTCCACCACCACCACCAGAAGAGGAATGGATTCCACCT CCACCACCGGAGAATGAACCAGCTCCTCCACCTCCTCCTGAGCCTGAAGAGCCTGCTGTG TCATTTGTTCATGCTGATACACTTCCTCAGCCATACGGAGGTCAAGCAAACTTGGGTTAT ATGCTTCCAGGAATGGAGTACTATCCCGCTTCTGGTACAGATGGCACAAATGCCAGCTAC TATATGCAAGCGAGCGATTCTCATATTCTTCAATCGCAGCAGCATTCTTACTACGCACCG TTGTCTGCAAGTGGCGTATCTATTCCTGTTGAGACCACATCGATCCCCCCAGTACCAGGT TCTTATTATAGCTATCCTTCTGTCACTATGGCTGCCACTGAAGTAGCGGCTGAATCTTCT GGATACTATGGTTCATCAACCTCTGCCATTTCTAGCGGTGAATTAGATAACAAAACAAGC TCAGCCTCCCTTGTTTCAAATAGTAATGTGAATCCTGTGGAGTCTGATAAAGTGATATCC AAGGAGCCTACAGTTGTTTCTTTGAGCCAATCAGTAGGGGCAGCCTCAGCTTCAGCACCA ACAGTACATGGTAGTTCTACTCTAGCTTCTACTAGTACCACTAATCAGACTAAAGTTCCT CGTACTAAGAAGCGACCTGTTGCTGTTGCATCATCACTGAGATCTAATAAGAAGGTCTCA AGTCTGGTGGATAAGTGGAAAGCTGCAAAAGAGGAGCTTTGTGATGAAGAGGAAGAGGAG CCTGAAGATGCTTTGGAGTACCTAGAAAGGAAACGTCGGAAGGAAATAGATGGGTGGCGT AAGCAACAAATAGCTAGTGGAGAAGCTAAGGAAAATGCTAATTTTGTTCCCCTTGGCGGT AACTGGCGTGACCGTGTAAAACGCAAAAGAGCTGAAGCAAAGAAGGAAGCAAAGACTGAA TCTGTTTGTGCAGCTGCCGAACAACACAAAGGAGAGCCTGATCTCTCAGAGCTTTCCAAG GGTCTCCCTTCTGGGTGGCAGGCATACATAGACGAGTCTACGAAGCAAGTGTACTATGGG AACAACCTCACTTCCGAGACTACTTGGGATCGGCCTAGCAAATAAAGATTGAACTAGTTG AAGATGGTCCAGTTTTGCTCTGTAGATATCCTCATACATCTATACGTTGCCTGTCTGGTT ATGAGGCTGTATTGTTTCTTTTATTTTCCACTGGGAGATCGTAGCAGGAAGGATACTTCA TTTTGGTATATAGAGGCATCCCTTGTAAGAGTGGCAACTTGAGATCGAATTACTGAAGCA AAAAATACTAGTTAGTTGCTGACCAAGGAAAAATGTGCCTTGTTAAGTGGTGGGAGTCTT TACCTCGATCTGATACCATTTGCTTTCAAAACAATGTCAATTATATTCGATTTTAGGTTG TGCTAAATTCTAGACTTCTCCAATTATTTAACATTAGAAAGAAAGATGATTTTTAAGTTT TTT
Protein Sequence (700 aa)
>BART1_0-u51812.003 700 150831_barley_pseudomolecules MEAVPDSSGMQIVGDIGGNWKAIMHEQSNQCYYWNTVTGETSWEIPDGLASGVAGDGVTS ASVPTHMGYSVEAQAHVLPHSNVEAYPSDVSVGNGTATYTAMGTYTHDAYAYTGAVASHE SVDIDPLQLAKYGEDLLQRLKLLERPHVAIDSLELIKREIEIRVADCNALSSYGSSLLPL WLHAEVHLKQLELSVSKFEASYTTRHGYLETVDAGHKAPNEAKVMAPSEGEHLKVDVSTL GIGTGDENIKVEEPCTTSSAQNSQCVAAAILRVESDSDEDMDVDMEVDEEGVEEHGGFTS MPNKEHPSSEQVRSPALSSLEDSAPPQQDNDIPPPPPPPEEEWIPPPPPENEPAPPPPPE PEEPAVSFVHADTLPQPYGGQANLGYMLPGMEYYPASGTDGTNASYYMQASDSHILQSQQ HSYYAPLSASGVSIPVETTSIPPVPGSYYSYPSVTMAATEVAAESSGYYGSSTSAISSGE LDNKTSSASLVSNSNVNPVESDKVISKEPTVVSLSQSVGAASASAPTVHGSSTLASTSTT NQTKVPRTKKRPVAVASSLRSNKKVSSLVDKWKAAKEELCDEEEEEPEDALEYLERKRRK EIDGWRKQQIASGEAKENANFVPLGGNWRDRVKRKRAEAKKEAKTESVCAAAEQHKGEPD LSELSKGLPSGWQAYIDESTKQVYYGNNLTSETTWDRPSK
