- Transcript BART1_0-u39718.002
- Transcript IDBART1_0-u39718.002
- Gene IDBART1_0-u39718
Exon Structure of BART1_0-u39718.002
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr5H | 1 | 615390084 | 615388469 | - |
| chr5H | 2 | 615390389 | 615390163 | - |
| chr5H | 3 | 615390625 | 615390551 | - |
| chr5H | 4 | 615390934 | 615390818 | - |
| chr5H | 5 | 615392416 | 615392324 | - |
| chr5H | 6 | 615394192 | 615394034 | - |
| chr5H | 7 | 615394470 | 615394336 | - |
| chr5H | 8 | 615394733 | 615394575 | - |
| chr5H | 9 | 615396770 | 615395625 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials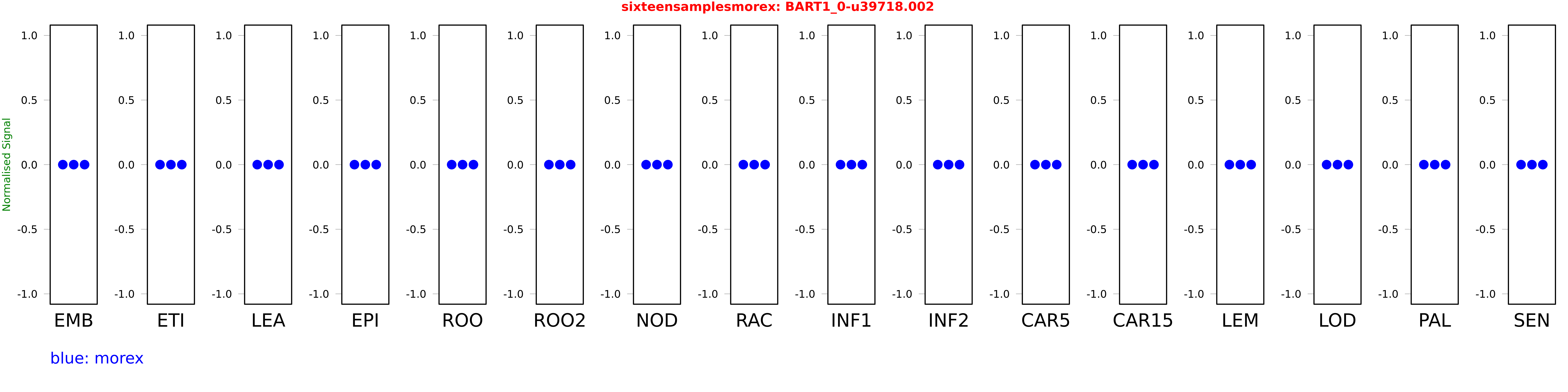
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0 | 0 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0 | 0 | 0 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os02g35150.1 | +1 | 2e-143 | 444 | 228/295 (77%) | protein|splicing factor U2AF, putative, expressed |
| TAIR PP10 | AT1G10320 @ ARAPORT AT1G10320.1 @ TAIR |
+1 | 2e-128 | 413 | 203/301 (67%) | Symbols: | Zinc finger C-x8-C-x5-C-x3-H type family protein | chr1:3384164-3388373 REVERSE LENGTH=757 |
| BRACH PP3 | Bradi3g45900.1.p | +1 | 0.0 | 626 | 334/397 (84%) | pacid=32822846 transcript=Bradi3g45900.1 locus=Bradi3g45900 ID=Bradi3g45900.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3727 bp)
>BART1_0-u39718.002 3727 150831_barley_pseudomolecules CCCAAACCTTTCTCAGCTTCACACAAACCAATCAATGCCCCGGAAATCACCAGAAATTTT ACCACACACAAGCACAACGAAGCAGCATACCGAACAACCGACTCGCCCTAAGCAGCCGAG GGCCCCAACCGGGGGAGGAATCGCGGGACGCGAGCTAGAGCAGAGCAGGTCGCGGGGGAG GGGAGGGGGGCAGAGCAGGGGCGGGGCTTGCCTTGGCGACGTAGTCCATCTCGGCGAACT TGAAGGCGATGCCGAGGCAGCCGAAGAGGACGGAGACGTTGGAGCAGGCGCGGGAGAAGT CCCCCGCCGCCATCGCGGGCTGCTGCTGCTGCTTGGCGATGGCCGCGAGCTGCTCGAACG ACGCGCCGATCCGCCGCAGCGGCTTGTCCGCCTCGTTGGACCCCATCGGCCGGTCTCCGC CGCCGCCGGCGTCCGCCGCGCTTGGAGGTTCTAGAGCTTGGGTGGCGTCGGCGTCTGGGC TGATGAGCTGTGCTCTTTTTTTTTCCCTCTCCCCCCCTCTCTCTGTGGTGAGATGCCTCG TTCTTTTGGTGGTTGTACGCGTTGTTGTGTGAAGAGGGCGACGGCGATGACGGCGAGGTT TTGGGGTCGGCCGGTCCTCTGATGGGCCTGAAAGTGGGCTCGGGCCATCTTGGACCATTC TCTTCTAGGCCCATGCGATTCTAAGAAAAACAAAATGCAAGGCCCATGAAACGTAAGTAC GTTTATTTGGGGGCCTTCGATAGACAAAGCCCTAAAACACTTTTCTGAAAGAAAAAAAAC CAAGAGGCCGAGGGGGTGGATCGAGGAGCTTCCGTCGGAGCGCCCATGTCCACGGCGGCC GCTGATCTCCCGGCGGCGTCTACGAGGAGCGAGAGGCGGAAGGAGCGGAAGAAGCAGCGC CGCCGGCGCGCGCGTAGGGAGGCGGCGGCGGCCGCGCGGGCTGCGGCAGAGGCGCTCGCC GCCGACCCAGAGGAGGGGCGCCGCCTCCGAGAGCTCGAGGAGGCCGAGGCCGACGCGTCC GACCGCGCCCGCCGCGCCTTTGAGGACGCCGAGCGCCGATGGCTCGAGAAGGCCGCCTCG CGTGCCGCCGAGGAGGCGGCTGCGGAAGAAGAGGCGTGTGCCGCCGAAGCTTCCACGCGC GATAAGTATAACGATGACCACCGAGACGAGACAGAAGAAGATGGGGAGTGGGAGTATGTT GAAGATGGACCAGCAGAGATCATATGGCAAGGAAATGAGATCATTGTGAAGAAGAAAAAG GTCAAAATACCAAAGAAGGCAGAGGCTAAACCACTAAGTCAAGAGGAAGAGAGACCTACA TCAAATCCGCTGCCACCTCAGTCTGTAGCTGTTGATTCTCAGAGGAGAGAACCTTCCTTG TCTGCTCGAGAACTACTTGAGAAAGTTGCTCAAGAGACTCCAAATTTCGGAACTGAACAG GATAAGGCCCATTGCCCATTTTACCTCAAAACAGCAGCTTGCCGCTTTGGAGTGCGCTGC AGCAGAGTTCACTTCTACCCTGACAAATCATGCACATTGCTCATGAAAAACATGTATAAT GGTCCGGGCCTTGTTTTGGAACAAGATGAAGGGCTTGAGTTTACAGACGAAGAGATTGAA CAGAGCTATGAAGAATTTTATGAAGACGTGCACACAGAATTTCTGAAATTTGGTGAACTT GTCAACTTCAAGGTATGTAGAAATGGATCGTTCCATCTTCGAGGAAATGTCTATGTGCAT TATAAAGCTTTGGATTCAGCTCTTCTTGCCTACAACAGCATGAATGGTCGATATTTTGCT GGGAAGCAGATAACATGCGAGTTTGTTGCTCTGACGAAATGGAAGTCTGCGATCTGTGGT GAGTACATGAGGTCAAGATTCAAGACATGTTCACATGGAGTTGCTTGTAATTTCATTCAT TGCTTTCGTAATCCTGGTGGAGATTATGAGTGGGCTGATTGGGACAATCCTCCACCTAAA TACTGGACTCGGAAGATGGCTGCTCTGTTTGGACCCTCAGATGATGCTGTTTTTGACAAG GCGAGTGATGCCCCAGATTTCGAACGGTTACACAGTTCAGACAGGAAGAGACTGAGAAGT TCTGATGGCAGGTATATTTCAAGTAGACATAGGGATGGAGATGCACATAAACGGCACTCA TCAAGAGTTTTTTCACATTCGAAGCAAGAACAGAGCAATTATATCATGAAATATGGGCAT ACTCGACATAGTAGAGAACCATCTACAGCAAAAAAGCAACGAGGCCATGAGATTGAAGAT GATATTGGCAGATACTGTTCGCCCGTGGAAAATGAAAGAGTGNNNNNNNNNNNNNNNNNN AAAGAGTGTCCCAAACACACAAGCGTGAAGAGAAACATAGAGATGGCCATGGCGATGGAG CACGTGGAGACAATGGTAAAATCAAGTCCAGGAAGCACAGATCTGAACAGCACGAAAGTC TGGAGTCTGGATGTTCTGATTGGCCTTCAGAATTTGCTGATGCAGGCACGAGCAGAAGTC CTTCATGCAGCAAGTCCACCAACAGGTACAGTAATCACAATAGGAGTCGGGGGCAATCCT TGGATGATTCCAATCTTGAAAGGGGCTACTCCACTGCGGACAAATCATCAGGGAAAGCGC ACATTGCCAAAAGTAGCTCGAGACATTACGTAGAAGATGACTCTTATGGTGAGAAAGGTA GTGGAAGAGACAAATCTGGAAAGCACAGTGATCATTGTGATGACCCTGATGATAGATGGC TAGCAACAACTAGTGACGTTGATTCAGATGGAGAAACTCAATTCCTGAGATCAAGCAGTG GAATTGGTAAAGGTGGAAAGAATGATAATGCTCCTCGAGACGAGGATGATCGTTGTCAAA GGTCCAGCAGTAGGTCTGCTAGGTCAAGAGCGAACGAGACGAAGAGCAGTAGGAAGAGGA AGAAGCATCACAGTGAAATCAGGAAGAATAGCGATAGTGATGACAGTCCGTCAGACTCTA GTGACATTAGGGATTCGAGCTCACATGCATGGAGGAGTCGATCAAGAAGCAGTGAGGAAG ATCTCTCTTTACACAGACGGAGGAAGGGTCGCCTAGGTCATGATTCGTGATGCTGCTGAA ATTTCTGATCATTTGGTCACTAGACTGTCGACTTCGGTTTCTGGACGATCAAATGTTGTA TAGATGCCAGTAGCTTATGCAGCAGTCCTTGCCAGCCTTGCTTTCTTCTTCACTTTGCCG ATAAGGTTCTTTTTCCGGATAGAGGTTTCTTGCAAAGCAGAAGGCTTGTAATAGTTTTGC TAATTTAGAGTTGTTTACATCTCACCATTGGTTCCGCCCTGGAATTCCGACTTTTTATGC CCATCACACTTGGTAGTAAGAGCATCTCCAACACATTTTCTGAAGTTACTCCCAATAATT CTGGGTATTGGTACGCTTGCAACTAGTCCCATCACCTAAGCTTAGCTCCCAATAATTTGT GTACATATACCTTGTTTTTTTATTCAAACCCTGCTGTTTCTAGTGCAAACATTTCAAATG CATAATATTATTTTTAATTCAAATTCAGGCCATGCACAAGAATTCAAACGGTAGGAGTGC TCCCTCCCTCTCTTCATCATCGCCCTCACCTCCTTCTTGTGCATCAATGGAAGATGGCAT GCTCAATACTGTCCTCTTTGGTGCTGTTTCTCAAATTACCATGCCACATTTCTCATTTTC CTTGACC
Protein Sequence (615 aa)
>BART1_0-u39718.002 615 150831_barley_pseudomolecules MSTAAADLPAASTRSERRKERKKQRRRRARREAAAAARAAAEALAADPEEGRRLRELEEA EADASDRARRAFEDAERRWLEKAASRAAEEAAAEEEACAAEASTRDKYNDDHRDETEEDG EWEYVEDGPAEIIWQGNEIIVKKKKVKIPKKAEAKPLSQEEERPTSNPLPPQSVAVDSQR REPSLSARELLEKVAQETPNFGTEQDKAHCPFYLKTAACRFGVRCSRVHFYPDKSCTLLM KNMYNGPGLVLEQDEGLEFTDEEIEQSYEEFYEDVHTEFLKFGELVNFKVCRNGSFHLRG NVYVHYKALDSALLAYNSMNGRYFAGKQITCEFVALTKWKSAICGEYMRSRFKTCSHGVA CNFIHCFRNPGGDYEWADWDNPPPKYWTRKMAALFGPSDDAVFDKASDAPDFERLHSSDR KRLRSSDGRYISSRHRDGDAHKRHSSRVFSHSKQEQSNYIMKYGHTRHSREPSTAKKQRG HEIEDDIGRYCSPVENERVXXXXXXKECPKHTSVKRNIEMAMAMEHVETMVKSSPGSTDL NSTKVWSLDVLIGLQNLLMQARAEVLHAASPPTGTVITIGVGGNPWMIPILKGATPLRTN HQGKRTLPKVARDIT
