- Transcript BART1_0-u39384.009
- Transcript IDBART1_0-u39384.009
- Gene IDBART1_0-u39384
Exon Structure of BART1_0-u39384.009
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr5H | 1 | 610332596 | 610332439 | + |
| chr5H | 2 | 610333705 | 610333396 | + |
| chr5H | 3 | 610335476 | 610335042 | + |
| chr5H | 4 | 610335745 | 610335608 | + |
| chr5H | 5 | 610335929 | 610335826 | + |
| chr5H | 6 | 610336235 | 610336014 | + |
| chr5H | 7 | 610337060 | 610336566 | + |
| chr5H | 8 | 610338426 | 610337999 | + |
| chr5H | 9 | 610338746 | 610338653 | + |
| chr5H | 10 | 610340032 | 610338820 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials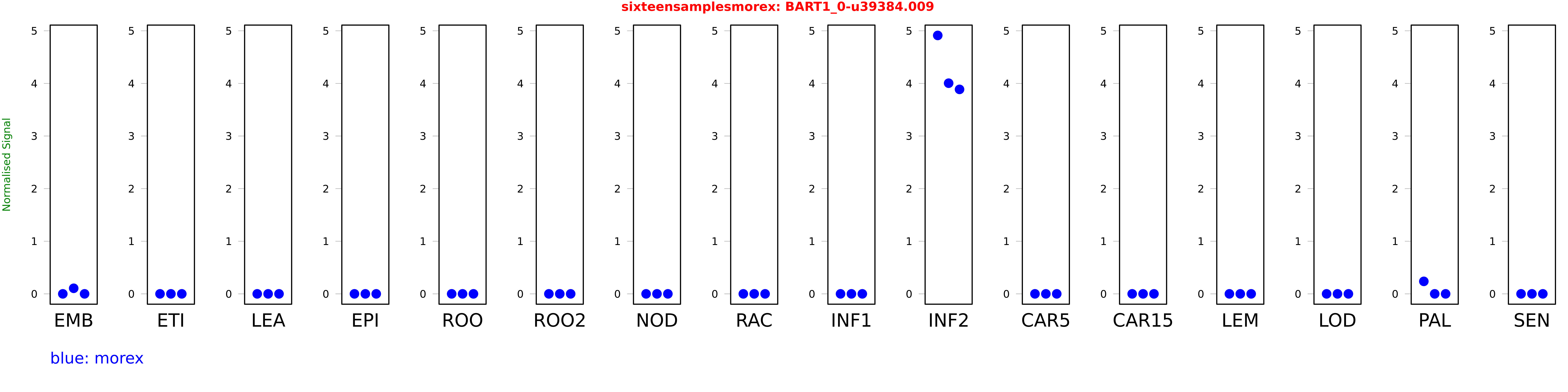
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0.105825 | 0 |
| morex | ETI | 0 | 0 | 0 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0 | 0 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 4.9121 | 4.00425 | 3.88655 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 0 | 0 | 0 |
| morex | PAL | 0.237476 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os05g22370.2 | +1 | 0.0 | 918 | 502/725 (69%) | protein|N-acetylglucosaminyl transferase component, putative, expressed |
| TAIR PP10 | AT3G57170 @ ARAPORT AT3G57170.1 @ TAIR |
+1 | 2e-116 | 374 | 233/531 (44%) | Symbols: | N-acetylglucosaminyl transferase component family protein / Gpi1 family protein | chr3:21159620-21162259 REVERSE LENGTH=560 |
| BRACH PP3 | Bradi1g06945.1.p | +1 | 0.0 | 1112 | 592/737 (80%) | pacid=32803293 transcript=Bradi1g06945.1 locus=Bradi1g06945 ID=Bradi1g06945.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3597 bp)
>BART1_0-u39384.009 3597 150831_barley_pseudomolecules CATCCCCCCGTGGCCTCCACGCAGCCGCCGGGTCGCTACCGCTCTCGCTGACTCGTGCAG GCACCGCGCAGGGGCTGCTGGCCGCTGCCTACTGCTCGGCTTTGGACAGGGGAAGCCCGG AGCCGTATCCCCACAGGGTATTTTTTTAATAGTTTTTGGCCACAGAAGATCAATCTCTGT TGAGTAAATAGGAATTTGGGCCTGTGATTCTGTGGTGCGGGTGCAGTTCCAGAACCTTGT GGGTCCAGCTGAACTCCCTATGCCCCACATTTAGTCTGCCCCTGCATGGACCGGTGCAGG ATCTGGTGGCCGCGCCAGCAACATCAGTCTGAATTGGAGTCCGTCTCTGCCAGATATCTC CTTTTTGGATGGCTCTTCCCCCATGCGGGGTCACTCGACATTGTTGTCGCGGCGTTCGTC TCCGAGGGAGAGATCTTGCGGTCTTTCCCCAACCTCGACACTTTCCAGACTGCTATCCTT TCCTCAAATAAAACGATGCCAACAGTGCTTCAAGAATCTGCAGCATTTACCATCCTTGGG GATTGTGTTGTACATCTTCCAAGAGATTTTGAAGGCTGCTGTGTTAAGCAAAAATATCAC CTGTTGCGGGCTCAAGTTGTTCAGGCAGAGCATGCTGACACAAACCAAGACAGTTCTATT GCATTGGGTGGATCATTGGGAAGCGAAGATCAAGACCAAAGTGAGAATAATGGAAAATGG GAATGTGATTGTAGTGTATTAGACGGTTTTCTGGATAAGTGCAAGAAGTCAGTAGTGAAA CGTGGCGACTGGGTGCACTTCTGCTGTAAGCCTGAAAAGAGCTTGAAAAGCAATCTGAAT CAAATTCCAGTGCTTCATCATTTGTATCTTGATGACCAGAAGGTTGAAATTAATCATTGC CACGTCATACTTTATGATGTTCCTGTTGCTGGTAGAAACCATTTCTCCCTGGGCGAGGAT GCACCTTATAGGATGAAGTCCCCCTTCAAAAGGCCAAATTGGATAAATAATCTGCAAAAG AAACGACCATTTCTTGATTTGGATCCGATTGTGTTGGCGCTTAATTGTTCAAATTCAGCT AGATTGTCAGTTGCTTGGAAAACTACTAATAACAGCTCTGCGGCTCATTTCTTGTTTGCA ACTGTGATTGATGCCTTAGTTCAAGTGGCACAACATTTCACAGGAATAATTTTGGCTTCA GTATCAACTATTATCTACATCTTCATTCAGCTGTTCCGAAAATGTTTGAGCCATGTCTCT GAGCACTTTATATTACAAAAGGTTTTCAGACACTCATGGAAAAACATGCATCTCCGTTGT TCCCAAATTCTTTATTGGCCGATTTTCCTCCAAGATACTTCCCTAAGCTCCTATGTGAAT GTCGAATATGCACACAAAGCTGCAATCCAGAAGCATGCTTTATGGTCAAATATTATCATG GATCTTCTGATGGGTTTCATCCTTGGAGCAGCACTATTGTTGAACATGGAGACTATTTGC TCTTGGATTTTTGCTCTTCTTCACTACATGACAGATGCTGTCTTGAGATCTGGTTGTGTG TGGCTGATGGGTGTTCCAGCAGGCTTTAAGCTGAATACTGAGTTAGCAGAGCTTCTAGGC ATGATTTCTTTGAATGCAATCCAAATTTATTCTACCCTTTGGTTCATGGTGGGAGGCTTC CTTAGGCATATAATTCAAGGCCTTGCATTTTCTGGAATTATTTTAGGTTTCACGGTTCCT GTTTCAATCTTCATTGACATTATCCAGCTTGCGACATTGCATGTTACCATGCTTCAGTGG TTAATCTCCTTAATATATTCAAGGCAGATTCAGACAGTGACATCATTGTGGCGTCTTTTC AGAGGGCGCAAGTGGAATCCTCTTAGGCAGAGATTAGATAGCTATGACTACACGGTTGAA CAACATGTGGTTGGTTCGCTGTTGTTTACACCAGTCTTGCTTCTTCTACCCACAGCTTCT ATATTCTACATATTTTTCTCTATCTTGAGCAGCACAATCATCTGTTTGTGTATAGTGTTG GAAACTGCAATATGTATAATCCACTCTACTCCTTATGCTGCGGTAATTCTATGGGTGACA AGGAGGCAAAGATTTCCTGCTGGATTAATGTTCCTTCCTATGTCATCGTCATCTGTATCT ACTGATGATGATGCTCTATCAGTTGAATATCATTCAACCAGTTTATCTGGCGAAAGGAAA ACAGACGAACTCATCCATGTACATTCAGTACCACTAGTTTCAGAACTTAACTGTAACTAT AACACCCTCGGACAAGTAATCGGGCCACATTACCAGAAAGTTTTCAACGGGATCGCTCTC CCCTTCTGCAAACAACTGGCACATGGAATCCTTAGGGGCACAAGGATACCTACAACACTG CATCTGCCCTCTTCTCCACTGCCCTGGATGCATATTGGCATTAGAGAATACTGGATGCTT TGCCGTCGCGCGACCAAGTGGGGAAGGAATTAAAAGTTCTCCGCTTATGATTAACTATTA GTCGCTCAGAGAATTTTTCTTAGTTGCCACCGTTTTGTAGTGGATGTTTTGCTTTAGTTG CCTAACACAGTAACAATTGTTGGTAGTTGCCAAGATGTTGCGTCCTGGACTGCTTTTGTA GTACTGATACCATTTGGAGTTGAGGAATCGACACGTAGGGAAAATTAATGTGTTTGTTAC AAGCATACAAGTTCTTCACATTCACGTAAATGCGTTGCGACCAAAATAGATGTGGTGGTG CACTCTCAGCGAGTAGGGTGGCATGACTCAGTACTTAAGTTTGGTGGTTGGTGTTATTTA GAGTTTGTTAGAACCTGTTCATGGTGGTGAAAATCTCAGATCGGTGTGATGGCCCGCTAG TAGAATTAAGGGAAATCACAGATCGGAATGTAACTTGGGATTGGAGAACGGGAACAACAG GAAAGAAATTATGGGATATGGAGAGGGTAGGTTGATAGTTCACATAATTCTCATTTCCTT CCAGGGGATGGCATCCAGCCTCTTTTAAACACACACCCACACACTTACAAGCGGGACCCC TGGTCCATAGACCACACACTACTACACCTGTCAGCTCACTCCATTGGTTCTTTATGTGGA CCCCTCTAGATCACCGCCTTCGTTATTGACACACCGAAATTTATTGGTCATGAAGGCTTG ACAATCAGGCTATCAATCTACCCTCTTGCATGATATATCTTTCTGATGCCTTTCAAACCC AAGTCTACGCATTTTCTCCTCCTTGAAACAAAAAAGGCAACACATTTTTCCTTGAATGTG TCGAGCCTAAACTTGTTTTACTCAGCTATAATTCGAGGAACTCTTTGTCATCTACTTTTT TTAATGGTTTTTCTCATCTACTTGGAACTCTTCATTGTATTCTCTTATCATACATACATT GGGTCAAGTATAAAATTGTATCACATATATAGGAGGTGCCCATGTATTAGAATGTGTGAG AGGAGAAGATATATGTGAAGCAGAACCAATGAAAAGAGATGGTCGTGCAGTACACTGGTA GTTTAATGCGCAACATTTCACATAATGAGTAGTGGTTGGTGCTGAGCTCGGCTTAAT
Protein Sequence (735 aa)
>BART1_0-u39384.009 735 150831_barley_pseudomolecules MDRCRIWWPRQQHQSELESVSARYLLFGWLFPHAGSLDIVVAAFVSEGEILRSFPNLDTF QTAILSSNKTMPTVLQESAAFTILGDCVVHLPRDFEGCCVKQKYHLLRAQVVQAEHADTN QDSSIALGGSLGSEDQDQSENNGKWECDCSVLDGFLDKCKKSVVKRGDWVHFCCKPEKSL KSNLNQIPVLHHLYLDDQKVEINHCHVILYDVPVAGRNHFSLGEDAPYRMKSPFKRPNWI NNLQKKRPFLDLDPIVLALNCSNSARLSVAWKTTNNSSAAHFLFATVIDALVQVAQHFTG IILASVSTIIYIFIQLFRKCLSHVSEHFILQKVFRHSWKNMHLRCSQILYWPIFLQDTSL SSYVNVEYAHKAAIQKHALWSNIIMDLLMGFILGAALLLNMETICSWIFALLHYMTDAVL RSGCVWLMGVPAGFKLNTELAELLGMISLNAIQIYSTLWFMVGGFLRHIIQGLAFSGIIL GFTVPVSIFIDIIQLATLHVTMLQWLISLIYSRQIQTVTSLWRLFRGRKWNPLRQRLDSY DYTVEQHVVGSLLFTPVLLLLPTASIFYIFFSILSSTIICLCIVLETAICIIHSTPYAAV ILWVTRRQRFPAGLMFLPMSSSSVSTDDDALSVEYHSTSLSGERKTDELIHVHSVPLVSE LNCNYNTLGQVIGPHYQKVFNGIALPFCKQLAHGILRGTRIPTTLHLPSSPLPWMHIGIR EYWMLCRRATKWGRN
