- Transcript BART1_0-u33637.022
- Transcript IDBART1_0-u33637.022
- Gene IDBART1_0-u33637
Exon Structure of BART1_0-u33637.022
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr5H | 1 | 84473806 | 84473684 | + |
| chr5H | 2 | 84474019 | 84473884 | + |
| chr5H | 3 | 84477710 | 84477550 | + |
| chr5H | 4 | 84479142 | 84478592 | + |
| chr5H | 5 | 84480117 | 84479843 | + |
| chr5H | 6 | 84480655 | 84480237 | + |
| chr5H | 7 | 84480831 | 84480728 | + |
| chr5H | 8 | 84483365 | 84483169 | + |
| chr5H | 9 | 84483994 | 84483785 | + |
| chr5H | 10 | 84490180 | 84489767 | + |
| chr5H | 11 | 84491799 | 84491518 | + |
| chr5H | 12 | 84493109 | 84492482 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials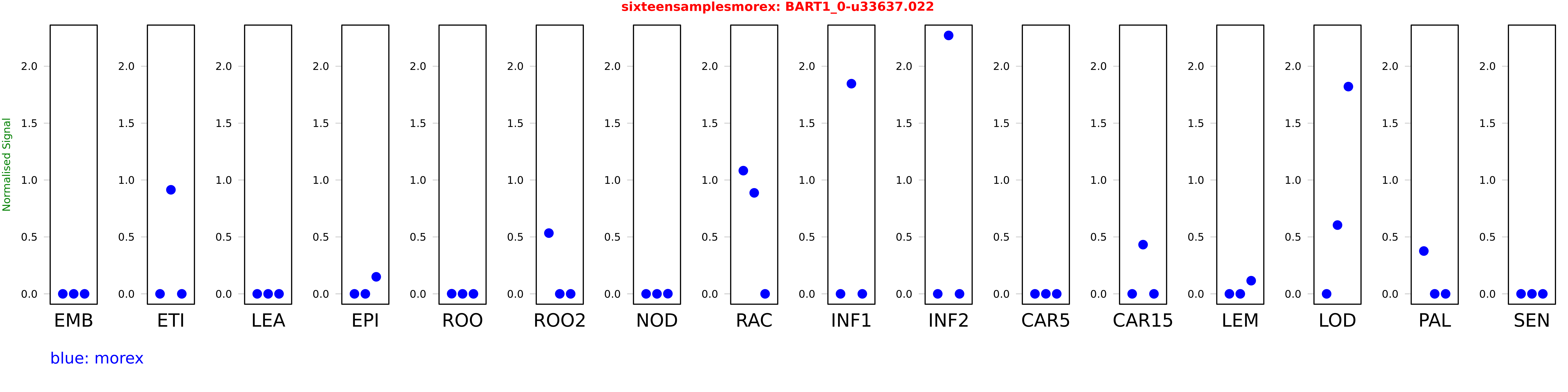
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0 |
| morex | ETI | 0 | 0.914162 | 0.00000000081719 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0.00000212864 | 0 | 0.149701 |
| morex | ROO | 0.00125892 | 0 | 0.0000451393 |
| morex | ROO2 | 0.533677 | 0 | 0 |
| morex | NOD | 0 | 0 | 0.00138546 |
| morex | RAC | 1.08237 | 0.887225 | 0 |
| morex | INF1 | 0 | 1.84641 | 0 |
| morex | INF2 | 0 | 2.27032 | 0 |
| morex | CAR5 | 0 | 0.000396307 | 0 |
| morex | CAR15 | 0 | 0.43232 | 0 |
| morex | LEM | 0 | 0 | 0.115645 |
| morex | LOD | 0 | 0.604103 | 1.82078 |
| morex | PAL | 0.37625 | 0 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os03g38740.1 | +2 | 0.0 | 1329 | 672/993 (68%) | protein|Dicer, putative, expressed |
| TAIR PP10 | AT3G03300 @ ARAPORT AT3G03300.3 @ TAIR |
+2 | 0.0 | 681 | 396/945 (42%) | Symbols: DCL2 | dicer-like 2 | chr3:768020-774833 REVERSE LENGTH=1388 |
| BRACH PP3 | Bradi1g15440.2.p | +2 | 0.0 | 1430 | 730/966 (76%) | pacid=32792174 transcript=Bradi1g15440.2 locus=Bradi1g15440 ID=Bradi1g15440.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3500 bp)
>BART1_0-u33637.022 3500 150831_barley_pseudomolecules TTAAAAGTCCTCAAGGCAAATCTACATGGATCGTCTCGGAAAAATGCGAAGAAAGGGATA TCAAAATTACACAAGGCATTCTTATATTGCACGGCTAATCTTGGAGTATGGCTAGCGGCA AAGGCTGCTGAGATACACTCAACAACTAACGAACAATTCTTATCTTTCTGGGGTGAAGAA CTGGATAAGAATGTTGAAGGCTTTGTTAGAAGCTATAGTGAAGAAGTATACAGGGAACTC TCATGTTTTTCAGAGAGAGGTACATATAATTGTGACCACACAAGTATTGGAGGAGGGGCT GAATGTGCCAGGTTGTCACTTAGTAATTCGCTTTGATCCACCAACGACTGGGCGCAGCTT TATACAGTCACGTGGTCGAGCTCGGATGCCAAACTCTGAATATGTTTTACTTGTTAGAAG GGGGGATGCAGAGGCCCTCTCTAAGACAGTGAAGTTCCTTGCAAGCGGGCAAATGATGCG GGAAGCATCTCTGAAGCTTGCATCTACTATGTGTCAACCACTAGAAAACACCTTGTTGCA GGGAGAATATTACCGTGTTGAATCTACTGGAGCAACCGTGACTATGAACTCAAGTGTCCA GTTGATTTATTTTTTCTGCTCGAAGCTCCCATCTGATGAGTCTGTTTTATGTACCTTAAA TTATGGTTTCCTTCTGTTTCATATCACAAATCATTTTCTGGCACAAGTATCTTATGTTTC TCACGGATAATATCTCTGTCTCAGATACTTTACACCTCTTCCGAGATTCATTATTGACAA GGAACTACGTACTTGCACCTTGTACCTTCCCAATAGCAGCCCTGTGCAGACTGTTAATGC AGAAGGAGAAGTTTCTGCCCTCAAAAAGGCAGTTTGTCTTAAGGCCTGCCGAGAATTACA CGCTGTCGGCGCCCTAACAGATTATCTGTTACCAGAATTTGGTTTTCCATGTGAAGAAGA ACCCGACATTGTTGTTGAGAAATACCAACACGAGCAGCCTGTGTACTTCCCAGAGGAATT TGTGTACAATTGGTCATCCTTTTCTCGTCTTGGAATATATTATTGCTATAAGATTTCAGT AGAAGGGTGCTTGAAAACAACTTATTGTCCAAATGACATACTTCTAGCTGTGAAATGTGA TTTGGGACCAGAGTTTGTCTCTACCTCACTTCAATTGTTTGGTGCGCAAGATAATGCTAG TCTTGCCATGAAATATGTGGGAATCATTCATCTGAATCAAGAACAGGTAGTTATGGCCAG AAGGTTTCAGACTACTATTCTTTCTTTGCTTATTAATAAAGACCATTCTGAGGTCAGTAA TGCTGTTAAATACTCCCATGAGATGCAAGTGTCCATTGGAATTGTCTACCTGCTGCTACC CTTAGTTTCTGGCAAAGTTGATTGGTGCAGTATCAAGTTCTCGACCTCGCAAGTGTATGT TGGCAGTAACAAGGATATCAGACATTGCCATTCTTGTAAACAAGTTGATTTACTGCAAAC AAAAGATGGTCCTCTGTGCAGATGCATGCTTGAAAGTTCCATTGTTTGCACCCCTCATAA TAGCAAATTCTATGCTGTTAATGGGTTCCTTGACTTAAATTCCAAATCTCTTCTGCACCT TCGTGATGGCAGTGCCCTTACCTACATAAACTATTTCAAGACTAGGCATGGACTCAGTCT AACACATGAAAATCAACCCCTGTTGGCAGCCAGGAGTCCTGTTGAAGTGCGGAATTTCCT CCACAAGCGCCATTATAAAAATAAAAAAGAATCGTGCACTTCACATGGTGTTGAGTTGCC ACCTGAGCTCTGCAGACTAGTCATGTCACCAGTCTCTACTAATACTTTGTACAGCTTCTC GATTATTCCTTCTGTAATGTACCGCATCCAGTGTCTGCTTCTTTCTGCAAAGTTGAAAGA TCAACTGGGCCCAATAATGCAGCAGTTTGCCATACCAGCTCTGAAGATTCTAGAAGCAGT AACAACCAAGGAATGCCAAGAGGAATTTTCACAGGAATCATTAGAAACCCTTGGGGATTC TTTCCTCAAGTACATCGCCACGCAACATCTGTATGTCAAACACAAGCTTCAGCATGAGGG GACATTAACCAAGATGAAGAAAAACTTAATCTCGAATGCAGCTCTATGCCAGCTGGCCTG CAACAACAATCTTGTGGGTTACATTCAAGGGGAAGAGTTTAACCCAAAAGGTTGGATCAT TCCAGGGCTTGGTTATGACATGTGTGGTAACAGCAAATTTTCCTTTTTAAGCTCCAATGA TATGCACACTTTGAGGAAAATGTCTGTTAGAAGTAAGAGAATAGCAGATACAGTTGAGGC ATTGATTGGAGCCTACCTTGGTGCTGCGGGAGAACAGGCTGCATTTGTTTTCCTAAAATC ATTAGGATTGGATATAGGATTTCACAGCACTATTCCACTTGAAAGGAAGATTGTATTAAA ATCTGAAAAGTTCATCAACGTGAGAAGTTTGGAAATGATTCTTGGTTATGAGTTCAAGGA TACTTCATTGTTGGTGGAAGCATTGACACATGGGTCATACCAGACTGCTGGCACTACTGC ATGCTATCAGAGGCTTGAGTTTCTCGGAGACGCTGTGTTGGACCATCTCTTCACTATATA TTTCTACAATAAGTACCCTGAATGTACTCCAGAATTGTTGACAGATCTTAGATCTGCTTC CGTTAACAACAATTGCTATGCGCATGCAGCTGCTAAAGCAGGACTAAACAAGCACATTCT TCATTCGTCATTGCCATTGCACGGAAGGATGGCTTACTACTTTGAGAATTTTACTAAGCA CCGATTTACAGGTCCATCACATGGTTGGGAAGCTGGCATTGGTCTACCCAAGGTTCTTGG GGATGTTATTGAATCATTAGCCGGTGCCATTTATCTTGACAGCAAGTATGACAAAGAGGT GGTTTGGAGAAGCATGAAAAAGCTATTGGAGCCTCTTGCGACACCGGAGACGGTTGAGCG TGACCCAGTGAAACTGCTGCAGGAATTTTGTGATCGCAGGTCTTATAGCAGGTCTTACAC AAAGGTCCAGAAAGATGGAGTGTCTTCGGTGGTTGGGGAGGTACAGGTAGAAGGGACTAC CTACTCAGCAACTGGAACTGGCCCTGATAAGGTTGTCGCAAAAAAGCTTGCAGCGAAGTT GCTGCTAGAAGATCTGAAGGCTATAGTACCATGACACGCTTGCATTTGCAGATGATGCAA TACTGTATAGCTCCTTCCGACTTCACTTGTATTGTGACTAGCCTGTAGGTAGCCTTTTGA AGTTTTTGTGGGAGTATGCGTGTACTTGACAAATGTGCAGCTCAATCATTGCTTGTGAAT ATTCAGACTATTGAACTGCACATCTGTGAAGTACGGGCATCGTCTCTCAGAACCATGGTA CAGACGATAATGCTACAGTATCCTTAGACTTCTACTTGTACAGTTGTAAGCACAATCTTG AGTGGAAAGCTGGAATATCT
Protein Sequence (668 aa)
>BART1_0-u33637.022 668 150831_barley_pseudomolecules MKYVGIIHLNQEQVVMARRFQTTILSLLINKDHSEVSNAVKYSHEMQVSIGIVYLLLPLV SGKVDWCSIKFSTSQVYVGSNKDIRHCHSCKQVDLLQTKDGPLCRCMLESSIVCTPHNSK FYAVNGFLDLNSKSLLHLRDGSALTYINYFKTRHGLSLTHENQPLLAARSPVEVRNFLHK RHYKNKKESCTSHGVELPPELCRLVMSPVSTNTLYSFSIIPSVMYRIQCLLLSAKLKDQL GPIMQQFAIPALKILEAVTTKECQEEFSQESLETLGDSFLKYIATQHLYVKHKLQHEGTL TKMKKNLISNAALCQLACNNNLVGYIQGEEFNPKGWIIPGLGYDMCGNSKFSFLSSNDMH TLRKMSVRSKRIADTVEALIGAYLGAAGEQAAFVFLKSLGLDIGFHSTIPLERKIVLKSE KFINVRSLEMILGYEFKDTSLLVEALTHGSYQTAGTTACYQRLEFLGDAVLDHLFTIYFY NKYPECTPELLTDLRSASVNNNCYAHAAAKAGLNKHILHSSLPLHGRMAYYFENFTKHRF TGPSHGWEAGIGLPKVLGDVIESLAGAIYLDSKYDKEVVWRSMKKLLEPLATPETVERDP VKLLQEFCDRRSYSRSYTKVQKDGVSSVVGEVQVEGTTYSATGTGPDKVVAKKLAAKLLL EDLKAIVP
