- Transcript BART1_0-u30899.020
- Transcript IDBART1_0-u30899.020
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.020
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593065664 | 593065502 | + |
| chr4H | 2 | 593068884 | 593066220 | + |
| chr4H | 3 | 593070199 | 593069549 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials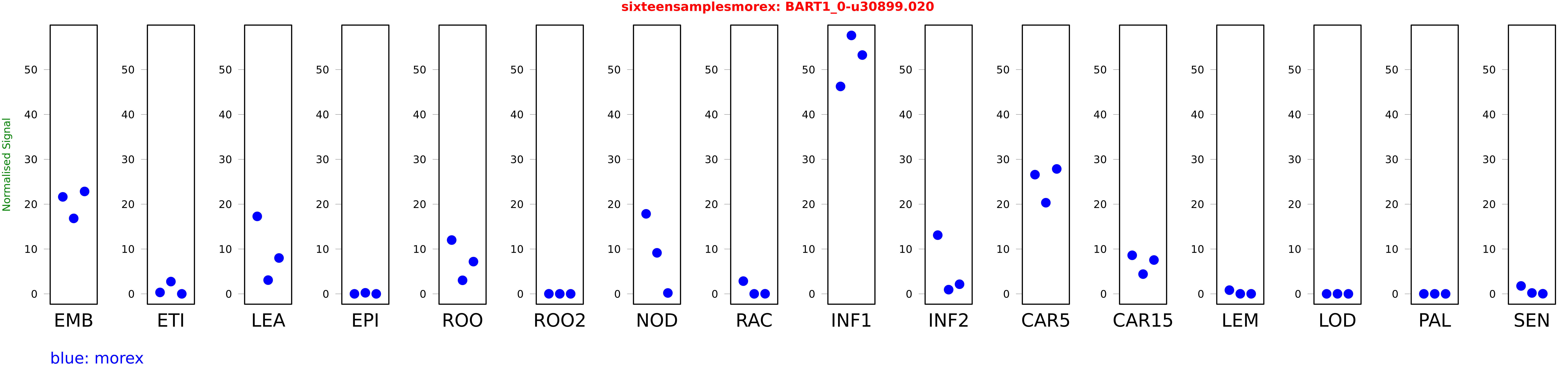
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 21.6252 | 16.8262 | 22.8349 |
| morex | ETI | 0.320923 | 2.7377 | 0.000440506 |
| morex | LEA | 17.2762 | 3.06377 | 7.995 |
| morex | EPI | 0.00015695 | 0.231486 | 0.000526763 |
| morex | ROO | 11.9958 | 3.0221 | 7.18671 |
| morex | ROO2 | 0.0135263 | 0.00328792 | 0.00269883 |
| morex | NOD | 17.8425 | 9.14598 | 0.195153 |
| morex | RAC | 2.83917 | 0.000206521 | 0.0188217 |
| morex | INF1 | 46.254 | 57.6239 | 53.2481 |
| morex | INF2 | 13.0897 | 0.934317 | 2.14588 |
| morex | CAR5 | 26.5795 | 20.3209 | 27.8548 |
| morex | CAR15 | 8.59989 | 4.40096 | 7.54246 |
| morex | LEM | 0.827941 | 0.0115908 | 0.00984228 |
| morex | LOD | 0.00254637 | 0.0110178 | 0.00000732105 |
| morex | PAL | 0.00025837 | 0.000050629 | 0.00000210004 |
| morex | SEN | 1.76551 | 0.199044 | 0.0284885 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +3 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+3 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +3 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3479 bp)
>BART1_0-u30899.020 3479 150831_barley_pseudomolecules CAGCTGTGTTTGTACCACCATCAATTATTATGTATTAGATTTGCCATTTTTCTGACTTGA TGTTATACAAACAGGAAGTTATTGTCTAATCTTAGTTCCGTTGTCGTACATATTCAAACA GCAAACAACAAACAACAAACATCTTCCCGATTCCTACTAATAGCTTTCTCCGAAGTATGG CTTGTGGACAAGTGTAGACCTGTAGACAAAGAGACAAAGAAATTGCTGACATAAGTCCTT ATTTGAGAAAGATCAGTTGTCAGCTTACAATGGCTGGAAATCAGCAGCCGGCTTCAGTTG TGTTGGATTATGCAGCAGTCCTCGACGCCAAACCCTTGCGCACACTGACCCCCATGTTCC CTGCACCGCTAGGGATGCACACTTTCACCCCTAAAAGCTCATCTTCAGTTGTCTGTGTCA CCCCGTTTGGACCATATGCTGGAGGTACCGAACTGGGAATGCCTGGTGGTGTTCCGCCAA TGTTAACATCACCGTCCGCTTCTGCAGATCCCAAGCAGGTGCAGCCATATACGGTTCATA TGAATGGAACTTCTAATGCTAATGGTACTACAAGCAACACAATGGTTACCCCTGTGTTGC AAACTCCTCCAGCAGTCACTACACAGGAGTCTGGTAAGAAGAAGAGGGGGATGGTTACCC CTGTGTTGCAAACTCCTCCAACAGTTACTACACAGGAGTCTGGTAAGAAGAAGAGGGGGA GGCCCAGGCGTGTGCAAGATACAACTACTGTTCCCCCGGTTCCTCCAGTTCAACCGGTTC CTACAGTTCATTCAGTTCCTTCAGCTCCTCCAGAAGTTAATAATATTGTTCTCCAGACAC CTACTTCAGCCGTCTCACAGGAATCTGGTAAGAGGAAGAGGGGACGACCCAAGCGTGTGC CAGATGTTTCTGTCTTATCAACTCCAGTGCCTGTAGCAGATGGTACGCCTATTTTACAGA CACCTCCTGCATCTAGTGTACATGAATCTGTTACAAAGAAAAGGGGGCGGCGATCCAAGC TTGTGCAGGATATTTCAGATACTTCAACTCCCCCAGTTCATTCAAAAGAGAGTGAGCCCT TTATGCAGACTCCTGCAGTCACCGTATTGGAGGATGGTAAGAGGAAGAGAGGGCGGCCGA AGCGTGTGCCTGATAGTTCAGTGACTCCTTCAAGTCATTCTGTCCTTACAGTAGATGTTG ACAGTGGTGACACATCAAAACGTGGCCGGCCTAGAAAAATTGACACAAGCCTATTGCACC TGCCATCTTTGTTTTCAGACGATCCCAGGGAATCCACAGATAATATACTTATGATGTTTG ATGCACTGCGACGGCGGCTCATGCAGCTGGATGAGGTGAAGCAAGTAGCAAAACAGCAGC AAAACTTGAAGGCTGGGAGTATCATGATGAGCGCTGAACTTCGCCTCAGTAAGAACAAGA GGATTGGAGAGGTTCCAGGTGTTGAGGTTGGTGACATGTTCTACTTCAGAATTGAGATGT GCCTGGTAGGGTTAAATAGTCAGAGCATGGCAGGGATAGATTATATGTCTGCTAAGTTTG GTAACGAGGAGGACCCCGTGGCCATTAGTATTGTGTCAGCTGGTGTGTATGAGGATGCTG AAGATAATGATCCAGATGTTCTGGTTTACAGTGGACATGGCATGTCTGGTAAGGATGACC AAAAGCTTGAGAGAGGTAATCTTGCACTGGAGAGAAGTTTGCATAGGGGTAATCCCATTA GAGTCGTTCGCACTGTGAAAGACTTGACTTGTTCAACTGGTAAGATATACATATATGATG GCCTTTACAGGATCAGAGAAGCCTGGGTAGAGAAAGGTAAATCTGGTTTCAATATGTTTA AACACAAGTTGCTCAGAGAACCTGGACAACCTGATGGCATTGCAGTGTGGAAGAAGACTG AAAAATGGAGGGAAAATCCATCCTCTAGAGATCGTGTTATAGTGCACGACATATCATACG GTGTGGAAAGTAAGCCTATCTGCCTTGTAAATGAGGTTGACGATGAGAAAGGTCCTAGCC ACTTCACGTATACAACTAAACTTAACTACATGAATTCCCCAAGCTCAATGAGAAAGATGC AAGGCTGCAAATGCACAAGCGTATGCCTACCCGGTGATAACAACTGTTCTTGTACGCATC GAAATGCTGGTGACCTTCCTTACAGTGCTTCGGGCATACTTGTTAGTCGGATGCCTATGT TATATGAGTGCAATGATTCATGCACTTGTTTACATAATTGCCGGAACCGGGTTGTACAAA AAGGTATCAAGATCCACTTTGAGGTGTTTAAAACGGGAGATCGAGGCTGGGGCCTGCGTA GTTGGGATCCAATCCGAGCAGGCACATTCATCTGTGAATATGCAGGTGTAATTGTTGACA AGAATGCTCTTGATGCAGAAGATGATTATATCTTTGAGACCCCTCCTTCGGAGCAGAACT TAAGATGGAACTATGCACCAGAATTACTGGGTGAACCTAGTCTTTCTGACTTAAATGAAT CATCTAAGCAGTTGCCAATAATTATTAGTGCAAAATATACTGGTAACATAGCTCGCTTCA TGAACCACAGCTGCTCACCTAATGTTTTTTGGCAACCAGTTCTGTATGACCATGGTGATG AGGGATATCCACACATCGCATTCTTTGCAATTAAGCATATTCCTCCAATGACAGAGCTTA CATATGATTATGGTCAGAGCGGTAATTCAGGGTGTCGGAGGTCTAAAAGTTGCTTATGTT GGTCTCGCAAGTGCAGGGGTTCCTTTGGCTAAATAATCAACCCTTAAGGGATACTTTCCT AGTTGAAGAAGCAGCTTATGGTGGCGGCGGCATTATGGGTGTGGTGTTCCAGCCACAGTT GAAATAACTTTGCTGCCTCTTTCTATCCGGGGTTGGAATGCGGGGTAGAGTACAAACGAC TAGATGTTGTAATAAAGGGCAAGCTATGCTTCTGTGGTGTTGTTAGTTGTAGTTGTCTGC TCCGGTTTAAGACCACCCAGTGGAATTCCTTCTATGTATTGTCATTGCGAAGATGTTTCA AGCCTGCAGTGAATCTATCGTTTTCGATTTGTAATCCTAGCGGATTAATGTGTCCTGTGA ATCTGTGATTGCCGTGGAGAGCGAATGAAGGCAACAATAATAATCCAGCTGGGCATTGTT CGTGCAGGGGTAAAGATAGGGGCAGAGTCAGTGCTGTGCGCTTGATTTATGATTCGGTGA ATGACTTTGATGGTTGATTGCTTAGATTCGCTGCTTTTGTTCTGTAGCGATCTTTATTTT CTTCTTATTCTAACCCTATTCTCTCTGAAATTTGTTTTTCTGTTCTTGTTCAGATTTGGT GTGAAGGATGGTGCTCTGTTCCTATGAGCAGTAGGCTCGCTGGGATTTTGGGCTTTGCCG TGTATTTTGGTCCCTGCTTGGCATACCGTTTTCTGCACATTACAGGTGTAAAATAAATA
Protein Sequence (840 aa)
>BART1_0-u30899.020 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
