- Transcript BART1_0-u30899.019
- Transcript IDBART1_0-u30899.019
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.019
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593065664 | 593065502 | + |
| chr4H | 2 | 593068884 | 593066220 | + |
| chr4H | 3 | 593070199 | 593069493 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials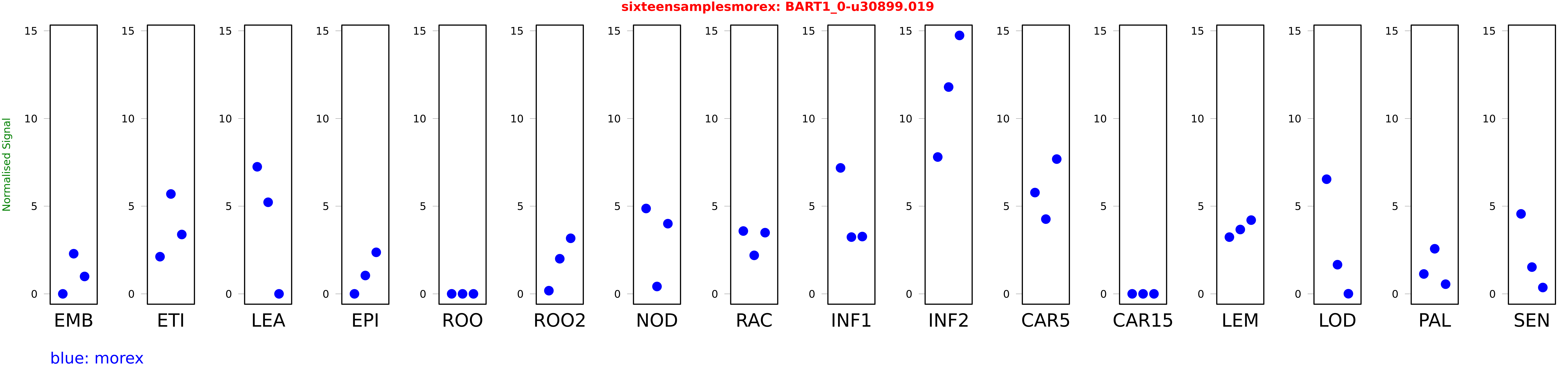
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.000025713 | 2.28945 | 0.992499 |
| morex | ETI | 2.11964 | 5.69568 | 3.38523 |
| morex | LEA | 7.24709 | 5.22316 | 0 |
| morex | EPI | 0.000556051 | 1.04689 | 2.36859 |
| morex | ROO | 0.00104281 | 0.0000000204811 | 0.000010144 |
| morex | ROO2 | 0.181357 | 2.00258 | 3.16911 |
| morex | NOD | 4.86851 | 0.420396 | 4.00276 |
| morex | RAC | 3.58309 | 2.19568 | 3.48942 |
| morex | INF1 | 7.18307 | 3.2368 | 3.26793 |
| morex | INF2 | 7.80398 | 11.7948 | 14.7384 |
| morex | CAR5 | 5.77625 | 4.26261 | 7.68579 |
| morex | CAR15 | 0.00103587 | 0 | 0.00000000227725 |
| morex | LEM | 3.23556 | 3.66986 | 4.20518 |
| morex | LOD | 6.53861 | 1.66325 | 0.00737465 |
| morex | PAL | 1.13376 | 2.57113 | 0.551608 |
| morex | SEN | 4.56025 | 1.52594 | 0.361695 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +3 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+3 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +3 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3535 bp)
>BART1_0-u30899.019 3535 150831_barley_pseudomolecules CAGCTGTGTTTGTACCACCATCAATTATTATGTATTAGATTTGCCATTTTTCTGACTTGA TGTTATACAAACAGGAAGTTATTGTCTAATCTTAGTTCCGTTGTCGTACATATTCAAACA GCAAACAACAAACAACAAACATCTTCCCGATTCCTACTAATAGCTTTCTCCGAAGTATGG CTTGTGGACAAGTGTAGACCTGTAGACAAAGAGACAAAGAAATTGCTGACATAAGTCCTT ATTTGAGAAAGATCAGTTGTCAGCTTACAATGGCTGGAAATCAGCAGCCGGCTTCAGTTG TGTTGGATTATGCAGCAGTCCTCGACGCCAAACCCTTGCGCACACTGACCCCCATGTTCC CTGCACCGCTAGGGATGCACACTTTCACCCCTAAAAGCTCATCTTCAGTTGTCTGTGTCA CCCCGTTTGGACCATATGCTGGAGGTACCGAACTGGGAATGCCTGGTGGTGTTCCGCCAA TGTTAACATCACCGTCCGCTTCTGCAGATCCCAAGCAGGTGCAGCCATATACGGTTCATA TGAATGGAACTTCTAATGCTAATGGTACTACAAGCAACACAATGGTTACCCCTGTGTTGC AAACTCCTCCAGCAGTCACTACACAGGAGTCTGGTAAGAAGAAGAGGGGGATGGTTACCC CTGTGTTGCAAACTCCTCCAACAGTTACTACACAGGAGTCTGGTAAGAAGAAGAGGGGGA GGCCCAGGCGTGTGCAAGATACAACTACTGTTCCCCCGGTTCCTCCAGTTCAACCGGTTC CTACAGTTCATTCAGTTCCTTCAGCTCCTCCAGAAGTTAATAATATTGTTCTCCAGACAC CTACTTCAGCCGTCTCACAGGAATCTGGTAAGAGGAAGAGGGGACGACCCAAGCGTGTGC CAGATGTTTCTGTCTTATCAACTCCAGTGCCTGTAGCAGATGGTACGCCTATTTTACAGA CACCTCCTGCATCTAGTGTACATGAATCTGTTACAAAGAAAAGGGGGCGGCGATCCAAGC TTGTGCAGGATATTTCAGATACTTCAACTCCCCCAGTTCATTCAAAAGAGAGTGAGCCCT TTATGCAGACTCCTGCAGTCACCGTATTGGAGGATGGTAAGAGGAAGAGAGGGCGGCCGA AGCGTGTGCCTGATAGTTCAGTGACTCCTTCAAGTCATTCTGTCCTTACAGTAGATGTTG ACAGTGGTGACACATCAAAACGTGGCCGGCCTAGAAAAATTGACACAAGCCTATTGCACC TGCCATCTTTGTTTTCAGACGATCCCAGGGAATCCACAGATAATATACTTATGATGTTTG ATGCACTGCGACGGCGGCTCATGCAGCTGGATGAGGTGAAGCAAGTAGCAAAACAGCAGC AAAACTTGAAGGCTGGGAGTATCATGATGAGCGCTGAACTTCGCCTCAGTAAGAACAAGA GGATTGGAGAGGTTCCAGGTGTTGAGGTTGGTGACATGTTCTACTTCAGAATTGAGATGT GCCTGGTAGGGTTAAATAGTCAGAGCATGGCAGGGATAGATTATATGTCTGCTAAGTTTG GTAACGAGGAGGACCCCGTGGCCATTAGTATTGTGTCAGCTGGTGTGTATGAGGATGCTG AAGATAATGATCCAGATGTTCTGGTTTACAGTGGACATGGCATGTCTGGTAAGGATGACC AAAAGCTTGAGAGAGGTAATCTTGCACTGGAGAGAAGTTTGCATAGGGGTAATCCCATTA GAGTCGTTCGCACTGTGAAAGACTTGACTTGTTCAACTGGTAAGATATACATATATGATG GCCTTTACAGGATCAGAGAAGCCTGGGTAGAGAAAGGTAAATCTGGTTTCAATATGTTTA AACACAAGTTGCTCAGAGAACCTGGACAACCTGATGGCATTGCAGTGTGGAAGAAGACTG AAAAATGGAGGGAAAATCCATCCTCTAGAGATCGTGTTATAGTGCACGACATATCATACG GTGTGGAAAGTAAGCCTATCTGCCTTGTAAATGAGGTTGACGATGAGAAAGGTCCTAGCC ACTTCACGTATACAACTAAACTTAACTACATGAATTCCCCAAGCTCAATGAGAAAGATGC AAGGCTGCAAATGCACAAGCGTATGCCTACCCGGTGATAACAACTGTTCTTGTACGCATC GAAATGCTGGTGACCTTCCTTACAGTGCTTCGGGCATACTTGTTAGTCGGATGCCTATGT TATATGAGTGCAATGATTCATGCACTTGTTTACATAATTGCCGGAACCGGGTTGTACAAA AAGGTATCAAGATCCACTTTGAGGTGTTTAAAACGGGAGATCGAGGCTGGGGCCTGCGTA GTTGGGATCCAATCCGAGCAGGCACATTCATCTGTGAATATGCAGGTGTAATTGTTGACA AGAATGCTCTTGATGCAGAAGATGATTATATCTTTGAGACCCCTCCTTCGGAGCAGAACT TAAGATGGAACTATGCACCAGAATTACTGGGTGAACCTAGTCTTTCTGACTTAAATGAAT CATCTAAGCAGTTGCCAATAATTATTAGTGCAAAATATACTGGTAACATAGCTCGCTTCA TGAACCACAGCTGCTCACCTAATGTTTTTTGGCAACCAGTTCTGTATGACCATGGTGATG AGGGATATCCACACATCGCATTCTTTGCAATTAAGCATATTCCTCCAATGACAGAGCTTA CATATGATTATGGTCAGAGCGGTAATTCAGGGTGTCGGAGGTCTAAAAGTTGCTTATGTT GGTCTCGCAAGTGCAGGGGTTCCTTTGGCTAAATAATCAACCCTTAAGGGATACTTTCCT AGTTGAAGATCCTGTTAAATGTATGCCTGGGTCGGAATCTGATGACCTTATCCTTGATGA ACAGAAGCAGCTTATGGTGGCGGCGGCATTATGGGTGTGGTGTTCCAGCCACAGTTGAAA TAACTTTGCTGCCTCTTTCTATCCGGGGTTGGAATGCGGGGTAGAGTACAAACGACTAGA TGTTGTAATAAAGGGCAAGCTATGCTTCTGTGGTGTTGTTAGTTGTAGTTGTCTGCTCCG GTTTAAGACCACCCAGTGGAATTCCTTCTATGTATTGTCATTGCGAAGATGTTTCAAGCC TGCAGTGAATCTATCGTTTTCGATTTGTAATCCTAGCGGATTAATGTGTCCTGTGAATCT GTGATTGCCGTGGAGAGCGAATGAAGGCAACAATAATAATCCAGCTGGGCATTGTTCGTG CAGGGGTAAAGATAGGGGCAGAGTCAGTGCTGTGCGCTTGATTTATGATTCGGTGAATGA CTTTGATGGTTGATTGCTTAGATTCGCTGCTTTTGTTCTGTAGCGATCTTTATTTTCTTC TTATTCTAACCCTATTCTCTCTGAAATTTGTTTTTCTGTTCTTGTTCAGATTTGGTGTGA AGGATGGTGCTCTGTTCCTATGAGCAGTAGGCTCGCTGGGATTTTGGGCTTTGCCGTGTA TTTTGGTCCCTGCTTGGCATACCGTTTTCTGCACATTACAGGTGTAAAATAAATA
Protein Sequence (840 aa)
>BART1_0-u30899.019 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
