- Transcript BART1_0-u30899.013
- Transcript IDBART1_0-u30899.013
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.013
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593062202 | 593062084 | + |
| chr4H | 2 | 593066130 | 593066072 | + |
| chr4H | 3 | 593068884 | 593066220 | + |
| chr4H | 4 | 593069956 | 593069549 | + |
| chr4H | 5 | 593070336 | 593070074 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials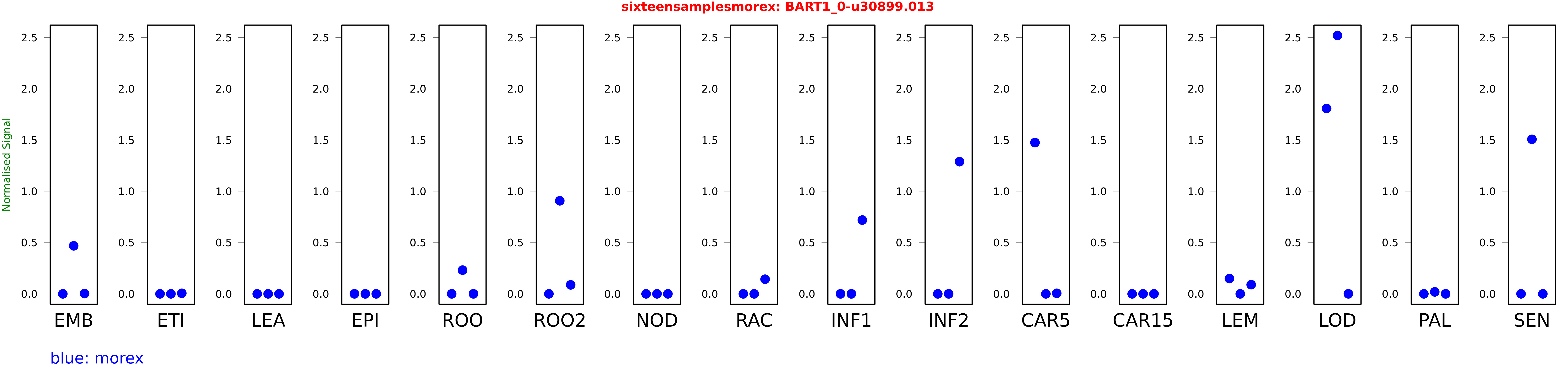
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0.468537 | 0.00236265 |
| morex | ETI | 0.000000000391418 | 0 | 0.00563084 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0.000152589 | 0.00000000382537 | 0.0000000860921 |
| morex | ROO | 0 | 0.2314 | 0 |
| morex | ROO2 | 0 | 0.908027 | 0.0876 |
| morex | NOD | 0 | 0 | 0 |
| morex | RAC | 0.00000000132986 | 0.0000000250713 | 0.142852 |
| morex | INF1 | 0.00000878028 | 0.00000874797 | 0.719891 |
| morex | INF2 | 0 | 0.0000027206 | 1.28912 |
| morex | CAR5 | 1.47585 | 0.0000000295998 | 0.00567045 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0.148884 | 0 | 0.089528 |
| morex | LOD | 1.80895 | 2.52118 | 0.000236953 |
| morex | PAL | 0.00000000144637 | 0.0193096 | 0.000000168087 |
| morex | SEN | 0.000208485 | 1.50766 | 0.00000327222 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +3 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+3 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +3 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3514 bp)
>BART1_0-u30899.013 3514 150831_barley_pseudomolecules CTGCCGTGGCGGCGCGGCGGCGCGATTCCTTTGCTGCTCGGCGGCAGCGGCCGACGGAAG CAGCGGCCGGGGTAGCAGCGACGGCAAGGCTCGGTTCTCTCCTCTCCCTCCCAACCAAGT AATCAGTGAACTGTGATTTGATGAATAATCAGACTTAAAAAAAAATCTAGATCTGCTGCT TTCTCCGAAGTATGGCTTGTGGACAAGTGTAGACCTGTAGACAAAGAGACAAAGAAATTG CTGACATAAGTCCTTATTTGAGAAAGATCAGTTGTCAGCTTACAATGGCTGGAAATCAGC AGCCGGCTTCAGTTGTGTTGGATTATGCAGCAGTCCTCGACGCCAAACCCTTGCGCACAC TGACCCCCATGTTCCCTGCACCGCTAGGGATGCACACTTTCACCCCTAAAAGCTCATCTT CAGTTGTCTGTGTCACCCCGTTTGGACCATATGCTGGAGGTACCGAACTGGGAATGCCTG GTGGTGTTCCGCCAATGTTAACATCACCGTCCGCTTCTGCAGATCCCAAGCAGGTGCAGC CATATACGGTTCATATGAATGGAACTTCTAATGCTAATGGTACTACAAGCAACACAATGG TTACCCCTGTGTTGCAAACTCCTCCAGCAGTCACTACACAGGAGTCTGGTAAGAAGAAGA GGGGGATGGTTACCCCTGTGTTGCAAACTCCTCCAACAGTTACTACACAGGAGTCTGGTA AGAAGAAGAGGGGGAGGCCCAGGCGTGTGCAAGATACAACTACTGTTCCCCCGGTTCCTC CAGTTCAACCGGTTCCTACAGTTCATTCAGTTCCTTCAGCTCCTCCAGAAGTTAATAATA TTGTTCTCCAGACACCTACTTCAGCCGTCTCACAGGAATCTGGTAAGAGGAAGAGGGGAC GACCCAAGCGTGTGCCAGATGTTTCTGTCTTATCAACTCCAGTGCCTGTAGCAGATGGTA CGCCTATTTTACAGACACCTCCTGCATCTAGTGTACATGAATCTGTTACAAAGAAAAGGG GGCGGCGATCCAAGCTTGTGCAGGATATTTCAGATACTTCAACTCCCCCAGTTCATTCAA AAGAGAGTGAGCCCTTTATGCAGACTCCTGCAGTCACCGTATTGGAGGATGGTAAGAGGA AGAGAGGGCGGCCGAAGCGTGTGCCTGATAGTTCAGTGACTCCTTCAAGTCATTCTGTCC TTACAGTAGATGTTGACAGTGGTGACACATCAAAACGTGGCCGGCCTAGAAAAATTGACA CAAGCCTATTGCACCTGCCATCTTTGTTTTCAGACGATCCCAGGGAATCCACAGATAATA TACTTATGATGTTTGATGCACTGCGACGGCGGCTCATGCAGCTGGATGAGGTGAAGCAAG TAGCAAAACAGCAGCAAAACTTGAAGGCTGGGAGTATCATGATGAGCGCTGAACTTCGCC TCAGTAAGAACAAGAGGATTGGAGAGGTTCCAGGTGTTGAGGTTGGTGACATGTTCTACT TCAGAATTGAGATGTGCCTGGTAGGGTTAAATAGTCAGAGCATGGCAGGGATAGATTATA TGTCTGCTAAGTTTGGTAACGAGGAGGACCCCGTGGCCATTAGTATTGTGTCAGCTGGTG TGTATGAGGATGCTGAAGATAATGATCCAGATGTTCTGGTTTACAGTGGACATGGCATGT CTGGTAAGGATGACCAAAAGCTTGAGAGAGGTAATCTTGCACTGGAGAGAAGTTTGCATA GGGGTAATCCCATTAGAGTCGTTCGCACTGTGAAAGACTTGACTTGTTCAACTGGTAAGA TATACATATATGATGGCCTTTACAGGATCAGAGAAGCCTGGGTAGAGAAAGGTAAATCTG GTTTCAATATGTTTAAACACAAGTTGCTCAGAGAACCTGGACAACCTGATGGCATTGCAG TGTGGAAGAAGACTGAAAAATGGAGGGAAAATCCATCCTCTAGAGATCGTGTTATAGTGC ACGACATATCATACGGTGTGGAAAGTAAGCCTATCTGCCTTGTAAATGAGGTTGACGATG AGAAAGGTCCTAGCCACTTCACGTATACAACTAAACTTAACTACATGAATTCCCCAAGCT CAATGAGAAAGATGCAAGGCTGCAAATGCACAAGCGTATGCCTACCCGGTGATAACAACT GTTCTTGTACGCATCGAAATGCTGGTGACCTTCCTTACAGTGCTTCGGGCATACTTGTTA GTCGGATGCCTATGTTATATGAGTGCAATGATTCATGCACTTGTTTACATAATTGCCGGA ACCGGGTTGTACAAAAAGGTATCAAGATCCACTTTGAGGTGTTTAAAACGGGAGATCGAG GCTGGGGCCTGCGTAGTTGGGATCCAATCCGAGCAGGCACATTCATCTGTGAATATGCAG GTGTAATTGTTGACAAGAATGCTCTTGATGCAGAAGATGATTATATCTTTGAGACCCCTC CTTCGGAGCAGAACTTAAGATGGAACTATGCACCAGAATTACTGGGTGAACCTAGTCTTT CTGACTTAAATGAATCATCTAAGCAGTTGCCAATAATTATTAGTGCAAAATATACTGGTA ACATAGCTCGCTTCATGAACCACAGCTGCTCACCTAATGTTTTTTGGCAACCAGTTCTGT ATGACCATGGTGATGAGGGATATCCACACATCGCATTCTTTGCAATTAAGCATATTCCTC CAATGACAGAGCTTACATATGATTATGGTCAGAGCGGTAATTCAGGGTGTCGGAGGTCTA AAAGTTGCTTATGTTGGTCTCGCAAGTGCAGGGGTTCCTTTGGCTAAATAATCAACCCTT AAGGGATACTTTCCTAGTTGAAGAAGCAGCTTATGGTGGCGGCGGCATTATGGGTGTGGT GTTCCAGCCACAGTTGAAATAACTTTGCTGCCTCTTTCTATCCGGGGTTGGAATGCGGGG TAGAGTACAAACGACTAGATGTTGTAATAAAGGGCAAGCTATGCTTCTGTGGTGTTGTTA GTTGTAGTTGTCTGCTCCGGTTTAAGACCACCCAGTGGAATTCCTTCTATGTATTGTCAT TGCGAAGATGTTTCAAGCCTGCAGTGAATCTATCGTTTTCGATTTGTAATCCTAGCGGAT TAATGTGTCCTGTGAATCTGTGATTGCCGTGGAGAGCGAATGAAGGCAACAATAATAATC CAGCTGGGCATTGTTCGTGCAGGGGTAAAGATAGGGGCAGAGTCAGTGCTGTGCGCTTGA TTTATGATTCGATTTGGTGTGAAGGATGGTGCTCTGTTCCTATGAGCAGTAGGCTCGCTG GGATTTTGGGCTTTGCCGTGTATTTTGGTCCCTGCTTGGCATACCGTTTTCTGCACATTA CAGGTGTAAAATAAATACAGACCTCCATCCACTGGTTTAGTTTCCAGCCGGAAGACACTC ATGTCCAGTAAAATACACCTGTAAATTTTACATCCCTGTATTAAATTACACCTATACCAA GCGCATCCTTAGCGTTTCATATATGAGAATCTGG
Protein Sequence (840 aa)
>BART1_0-u30899.013 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
