- Transcript BART1_0-u30899.005
- Transcript IDBART1_0-u30899.005
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.005
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593062202 | 593062069 | + |
| chr4H | 2 | 593068884 | 593066220 | + |
| chr4H | 3 | 593070616 | 593070074 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials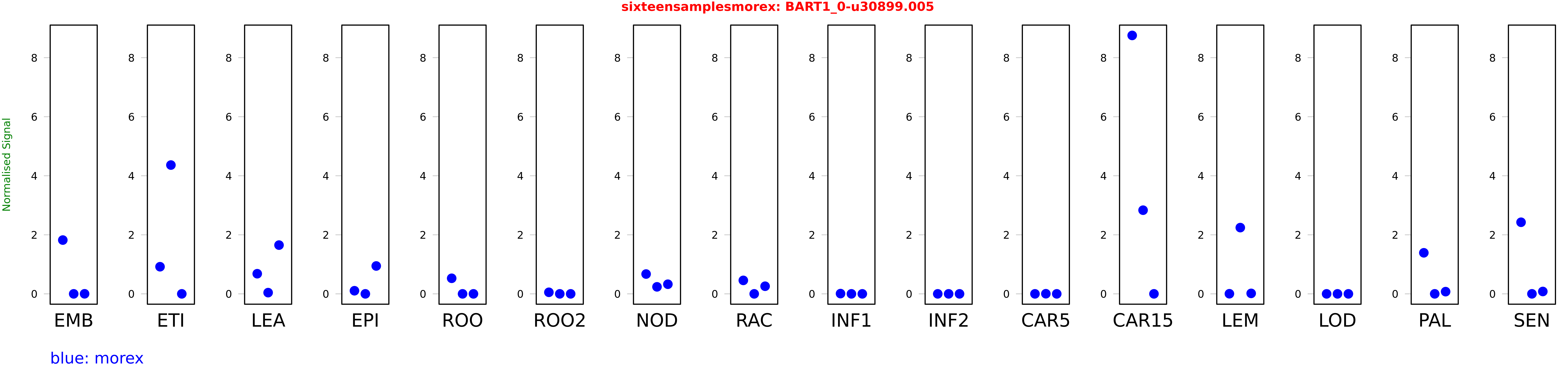
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 1.82228 | 0 | 0.00174656 |
| morex | ETI | 0.919067 | 4.36341 | 0.0000103449 |
| morex | LEA | 0.682239 | 0.0413035 | 1.65175 |
| morex | EPI | 0.105702 | 0.000147071 | 0.944596 |
| morex | ROO | 0.527514 | 0.00000185954 | 0 |
| morex | ROO2 | 0.0528625 | 0.0000126953 | 0 |
| morex | NOD | 0.671398 | 0.237596 | 0.3264 |
| morex | RAC | 0.456555 | 0.0000369779 | 0.25838 |
| morex | INF1 | 0.00861871 | 0.00000303361 | 0 |
| morex | INF2 | 0 | 0.00000682615 | 0.000000257205 |
| morex | CAR5 | 0.0000113802 | 0.00417293 | 0 |
| morex | CAR15 | 8.75522 | 2.8345 | 0.000152155 |
| morex | LEM | 0.00370277 | 2.24425 | 0.0158416 |
| morex | LOD | 0 | 0.00000205094 | 0.00000107026 |
| morex | PAL | 1.39069 | 0.000343469 | 0.0748371 |
| morex | SEN | 2.42581 | 0.00043645 | 0.0803636 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +1 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+1 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +1 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3342 bp)
>BART1_0-u30899.005 3342 150831_barley_pseudomolecules CCCCCCCCCCCCCCCCTGCCGTGGCGGCGCGGCGGCGCGATTCCTTTGCTGCTCGGCGGC AGCGGCCGACGGAAGCAGCGGCCGGGGTAGCAGCGACGGCAAGGCTCGGTTCTCTCCTCT CCCTCCCAACCAAGCTTTCTCCGAAGTATGGCTTGTGGACAAGTGTAGACCTGTAGACAA AGAGACAAAGAAATTGCTGACATAAGTCCTTATTTGAGAAAGATCAGTTGTCAGCTTACA ATGGCTGGAAATCAGCAGCCGGCTTCAGTTGTGTTGGATTATGCAGCAGTCCTCGACGCC AAACCCTTGCGCACACTGACCCCCATGTTCCCTGCACCGCTAGGGATGCACACTTTCACC CCTAAAAGCTCATCTTCAGTTGTCTGTGTCACCCCGTTTGGACCATATGCTGGAGGTACC GAACTGGGAATGCCTGGTGGTGTTCCGCCAATGTTAACATCACCGTCCGCTTCTGCAGAT CCCAAGCAGGTGCAGCCATATACGGTTCATATGAATGGAACTTCTAATGCTAATGGTACT ACAAGCAACACAATGGTTACCCCTGTGTTGCAAACTCCTCCAGCAGTCACTACACAGGAG TCTGGTAAGAAGAAGAGGGGGATGGTTACCCCTGTGTTGCAAACTCCTCCAACAGTTACT ACACAGGAGTCTGGTAAGAAGAAGAGGGGGAGGCCCAGGCGTGTGCAAGATACAACTACT GTTCCCCCGGTTCCTCCAGTTCAACCGGTTCCTACAGTTCATTCAGTTCCTTCAGCTCCT CCAGAAGTTAATAATATTGTTCTCCAGACACCTACTTCAGCCGTCTCACAGGAATCTGGT AAGAGGAAGAGGGGACGACCCAAGCGTGTGCCAGATGTTTCTGTCTTATCAACTCCAGTG CCTGTAGCAGATGGTACGCCTATTTTACAGACACCTCCTGCATCTAGTGTACATGAATCT GTTACAAAGAAAAGGGGGCGGCGATCCAAGCTTGTGCAGGATATTTCAGATACTTCAACT CCCCCAGTTCATTCAAAAGAGAGTGAGCCCTTTATGCAGACTCCTGCAGTCACCGTATTG GAGGATGGTAAGAGGAAGAGAGGGCGGCCGAAGCGTGTGCCTGATAGTTCAGTGACTCCT TCAAGTCATTCTGTCCTTACAGTAGATGTTGACAGTGGTGACACATCAAAACGTGGCCGG CCTAGAAAAATTGACACAAGCCTATTGCACCTGCCATCTTTGTTTTCAGACGATCCCAGG GAATCCACAGATAATATACTTATGATGTTTGATGCACTGCGACGGCGGCTCATGCAGCTG GATGAGGTGAAGCAAGTAGCAAAACAGCAGCAAAACTTGAAGGCTGGGAGTATCATGATG AGCGCTGAACTTCGCCTCAGTAAGAACAAGAGGATTGGAGAGGTTCCAGGTGTTGAGGTT GGTGACATGTTCTACTTCAGAATTGAGATGTGCCTGGTAGGGTTAAATAGTCAGAGCATG GCAGGGATAGATTATATGTCTGCTAAGTTTGGTAACGAGGAGGACCCCGTGGCCATTAGT ATTGTGTCAGCTGGTGTGTATGAGGATGCTGAAGATAATGATCCAGATGTTCTGGTTTAC AGTGGACATGGCATGTCTGGTAAGGATGACCAAAAGCTTGAGAGAGGTAATCTTGCACTG GAGAGAAGTTTGCATAGGGGTAATCCCATTAGAGTCGTTCGCACTGTGAAAGACTTGACT TGTTCAACTGGTAAGATATACATATATGATGGCCTTTACAGGATCAGAGAAGCCTGGGTA GAGAAAGGTAAATCTGGTTTCAATATGTTTAAACACAAGTTGCTCAGAGAACCTGGACAA CCTGATGGCATTGCAGTGTGGAAGAAGACTGAAAAATGGAGGGAAAATCCATCCTCTAGA GATCGTGTTATAGTGCACGACATATCATACGGTGTGGAAAGTAAGCCTATCTGCCTTGTA AATGAGGTTGACGATGAGAAAGGTCCTAGCCACTTCACGTATACAACTAAACTTAACTAC ATGAATTCCCCAAGCTCAATGAGAAAGATGCAAGGCTGCAAATGCACAAGCGTATGCCTA CCCGGTGATAACAACTGTTCTTGTACGCATCGAAATGCTGGTGACCTTCCTTACAGTGCT TCGGGCATACTTGTTAGTCGGATGCCTATGTTATATGAGTGCAATGATTCATGCACTTGT TTACATAATTGCCGGAACCGGGTTGTACAAAAAGGTATCAAGATCCACTTTGAGGTGTTT AAAACGGGAGATCGAGGCTGGGGCCTGCGTAGTTGGGATCCAATCCGAGCAGGCACATTC ATCTGTGAATATGCAGGTGTAATTGTTGACAAGAATGCTCTTGATGCAGAAGATGATTAT ATCTTTGAGACCCCTCCTTCGGAGCAGAACTTAAGATGGAACTATGCACCAGAATTACTG GGTGAACCTAGTCTTTCTGACTTAAATGAATCATCTAAGCAGTTGCCAATAATTATTAGT GCAAAATATACTGGTAACATAGCTCGCTTCATGAACCACAGCTGCTCACCTAATGTTTTT TGGCAACCAGTTCTGTATGACCATGGTGATGAGGGATATCCACACATCGCATTCTTTGCA ATTAAGCATATTCCTCCAATGACAGAGCTTACATATGATTATGGTCAGAGCGGTAATTCA GGGTGTCGGAGGTCTAAAAGTTGCTTATGTTGGTCTCGCAAGTGCAGGGGTTCCTTTGGC TAAATAATCAACCCTTAAGGGATACTTTCCTAGTTGAAGATTTGGTGTGAAGGATGGTGC TCTGTTCCTATGAGCAGTAGGCTCGCTGGGATTTTGGGCTTTGCCGTGTATTTTGGTCCC TGCTTGGCATACCGTTTTCTGCACATTACAGGTGTAAAATAAATACAGACCTCCATCCAC TGGTTTAGTTTCCAGCCGGAAGACACTCATGTCCAGTAAAATACACCTGTAAATTTTACA TCCCTGTATTAAATTACACCTATACCAAGCGCATCCTTAGCGTTTCATATATGAGAATCT GGTGGTTACCCTATAGTCTGTCTGGACAGCATGCATGAAATTCATTCATGAAGGAATCAA CCAAAGCTCTCAAGCGACGCATATGTTCATCTAGCATTTCATATCCGAACTTGATATGAT ACATGAGAATAAGGCCCAATTCTCTCAGAATCTAGGGATTCTCTCATAATCAAATCCCTT CCCAGCTTCTACAAGAAACTTTAAGCTCCATTTGTTTTACTCTCTCCGTCTCGGTTTACA AGTCCGGCATATGTACCTAGGTCGTCAATTTGATCAACTTAA
Protein Sequence (840 aa)
>BART1_0-u30899.005 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
