- Transcript BART1_0-u30899.004
- Transcript IDBART1_0-u30899.004
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.004
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593062202 | 593062067 | + |
| chr4H | 2 | 593066130 | 593066072 | + |
| chr4H | 3 | 593068884 | 593066220 | + |
| chr4H | 4 | 593070412 | 593070074 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials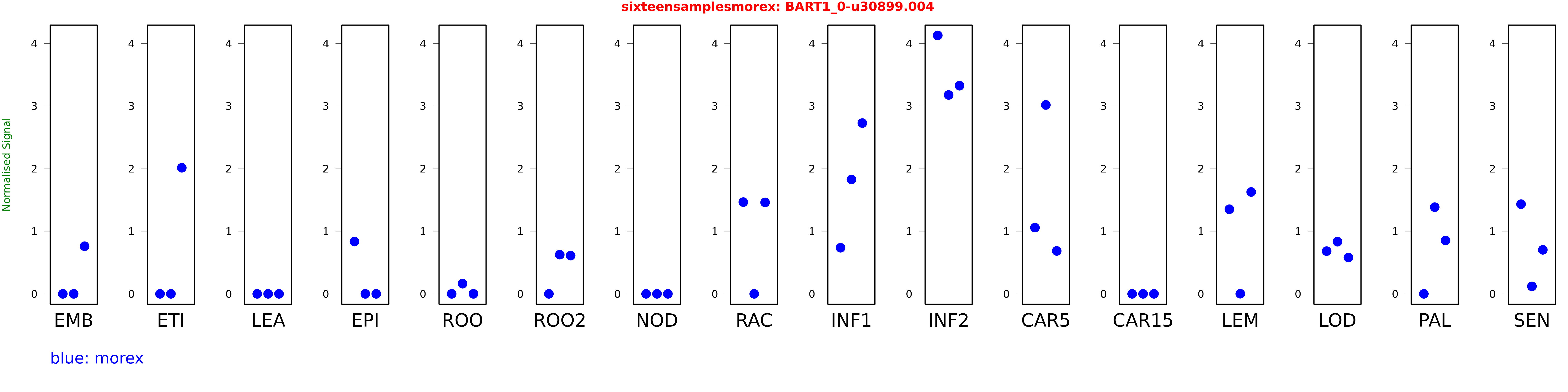
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 0 | 0.760925 |
| morex | ETI | 0.0000000878912 | 0.00000563562 | 2.01409 |
| morex | LEA | 0 | 0 | 0.00000000154178 |
| morex | EPI | 0.835012 | 0.000000135153 | 0.0000121564 |
| morex | ROO | 0 | 0.16205 | 0 |
| morex | ROO2 | 0.00000090766 | 0.625964 | 0.610583 |
| morex | NOD | 0 | 0.0000000499758 | 0 |
| morex | RAC | 1.46557 | 0.0000000975116 | 1.46035 |
| morex | INF1 | 0.736283 | 1.82759 | 2.7282 |
| morex | INF2 | 4.12827 | 3.17707 | 3.32441 |
| morex | CAR5 | 1.05808 | 3.01762 | 0.685643 |
| morex | CAR15 | 0 | 0.0000000554991 | 0 |
| morex | LEM | 1.35181 | 0.00200188 | 1.62739 |
| morex | LOD | 0.682418 | 0.833119 | 0.580257 |
| morex | PAL | 0.00000427194 | 1.38481 | 0.852592 |
| morex | SEN | 1.43285 | 0.119761 | 0.703518 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +2 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+2 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +2 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3199 bp)
>BART1_0-u30899.004 3199 150831_barley_pseudomolecules NNCCCCCCCCCCCCCCCCTGCCGTGGCGGCGCGGCGGCGCGATTCCTTTGCTGCTCGGCG GCAGCGGCCGACGGAAGCAGCGGCCGGGGTAGCAGCGACGGCAAGGCTCGGTTCTCTCCT CTCCCTCCCAACCAAGTAATCAGTGAACTGTGATTTGATGAATAATCAGACTTAAAAAAA AATCTAGATCTGCTGCTTTCTCCGAAGTATGGCTTGTGGACAAGTGTAGACCTGTAGACA AAGAGACAAAGAAATTGCTGACATAAGTCCTTATTTGAGAAAGATCAGTTGTCAGCTTAC AATGGCTGGAAATCAGCAGCCGGCTTCAGTTGTGTTGGATTATGCAGCAGTCCTCGACGC CAAACCCTTGCGCACACTGACCCCCATGTTCCCTGCACCGCTAGGGATGCACACTTTCAC CCCTAAAAGCTCATCTTCAGTTGTCTGTGTCACCCCGTTTGGACCATATGCTGGAGGTAC CGAACTGGGAATGCCTGGTGGTGTTCCGCCAATGTTAACATCACCGTCCGCTTCTGCAGA TCCCAAGCAGGTGCAGCCATATACGGTTCATATGAATGGAACTTCTAATGCTAATGGTAC TACAAGCAACACAATGGTTACCCCTGTGTTGCAAACTCCTCCAGCAGTCACTACACAGGA GTCTGGTAAGAAGAAGAGGGGGATGGTTACCCCTGTGTTGCAAACTCCTCCAACAGTTAC TACACAGGAGTCTGGTAAGAAGAAGAGGGGGAGGCCCAGGCGTGTGCAAGATACAACTAC TGTTCCCCCGGTTCCTCCAGTTCAACCGGTTCCTACAGTTCATTCAGTTCCTTCAGCTCC TCCAGAAGTTAATAATATTGTTCTCCAGACACCTACTTCAGCCGTCTCACAGGAATCTGG TAAGAGGAAGAGGGGACGACCCAAGCGTGTGCCAGATGTTTCTGTCTTATCAACTCCAGT GCCTGTAGCAGATGGTACGCCTATTTTACAGACACCTCCTGCATCTAGTGTACATGAATC TGTTACAAAGAAAAGGGGGCGGCGATCCAAGCTTGTGCAGGATATTTCAGATACTTCAAC TCCCCCAGTTCATTCAAAAGAGAGTGAGCCCTTTATGCAGACTCCTGCAGTCACCGTATT GGAGGATGGTAAGAGGAAGAGAGGGCGGCCGAAGCGTGTGCCTGATAGTTCAGTGACTCC TTCAAGTCATTCTGTCCTTACAGTAGATGTTGACAGTGGTGACACATCAAAACGTGGCCG GCCTAGAAAAATTGACACAAGCCTATTGCACCTGCCATCTTTGTTTTCAGACGATCCCAG GGAATCCACAGATAATATACTTATGATGTTTGATGCACTGCGACGGCGGCTCATGCAGCT GGATGAGGTGAAGCAAGTAGCAAAACAGCAGCAAAACTTGAAGGCTGGGAGTATCATGAT GAGCGCTGAACTTCGCCTCAGTAAGAACAAGAGGATTGGAGAGGTTCCAGGTGTTGAGGT TGGTGACATGTTCTACTTCAGAATTGAGATGTGCCTGGTAGGGTTAAATAGTCAGAGCAT GGCAGGGATAGATTATATGTCTGCTAAGTTTGGTAACGAGGAGGACCCCGTGGCCATTAG TATTGTGTCAGCTGGTGTGTATGAGGATGCTGAAGATAATGATCCAGATGTTCTGGTTTA CAGTGGACATGGCATGTCTGGTAAGGATGACCAAAAGCTTGAGAGAGGTAATCTTGCACT GGAGAGAAGTTTGCATAGGGGTAATCCCATTAGAGTCGTTCGCACTGTGAAAGACTTGAC TTGTTCAACTGGTAAGATATACATATATGATGGCCTTTACAGGATCAGAGAAGCCTGGGT AGAGAAAGGTAAATCTGGTTTCAATATGTTTAAACACAAGTTGCTCAGAGAACCTGGACA ACCTGATGGCATTGCAGTGTGGAAGAAGACTGAAAAATGGAGGGAAAATCCATCCTCTAG AGATCGTGTTATAGTGCACGACATATCATACGGTGTGGAAAGTAAGCCTATCTGCCTTGT AAATGAGGTTGACGATGAGAAAGGTCCTAGCCACTTCACGTATACAACTAAACTTAACTA CATGAATTCCCCAAGCTCAATGAGAAAGATGCAAGGCTGCAAATGCACAAGCGTATGCCT ACCCGGTGATAACAACTGTTCTTGTACGCATCGAAATGCTGGTGACCTTCCTTACAGTGC TTCGGGCATACTTGTTAGTCGGATGCCTATGTTATATGAGTGCAATGATTCATGCACTTG TTTACATAATTGCCGGAACCGGGTTGTACAAAAAGGTATCAAGATCCACTTTGAGGTGTT TAAAACGGGAGATCGAGGCTGGGGCCTGCGTAGTTGGGATCCAATCCGAGCAGGCACATT CATCTGTGAATATGCAGGTGTAATTGTTGACAAGAATGCTCTTGATGCAGAAGATGATTA TATCTTTGAGACCCCTCCTTCGGAGCAGAACTTAAGATGGAACTATGCACCAGAATTACT GGGTGAACCTAGTCTTTCTGACTTAAATGAATCATCTAAGCAGTTGCCAATAATTATTAG TGCAAAATATACTGGTAACATAGCTCGCTTCATGAACCACAGCTGCTCACCTAATGTTTT TTGGCAACCAGTTCTGTATGACCATGGTGATGAGGGATATCCACACATCGCATTCTTTGC AATTAAGCATATTCCTCCAATGACAGAGCTTACATATGATTATGGTCAGAGCGGTAATTC AGGGTGTCGGAGGTCTAAAAGTTGCTTATGTTGGTCTCGCAAGTGCAGGGGTTCCTTTGG CTAAATAATCAACCCTTAAGGGATACTTTCCTAGTTGAAGATTTGGTGTGAAGGATGGTG CTCTGTTCCTATGAGCAGTAGGCTCGCTGGGATTTTGGGCTTTGCCGTGTATTTTGGTCC CTGCTTGGCATACCGTTTTCTGCACATTACAGGTGTAAAATAAATACAGACCTCCATCCA CTGGTTTAGTTTCCAGCCGGAAGACACTCATGTCCAGTAAAATACACCTGTAAATTTTAC ATCCCTGTATTAAATTACACCTATACCAAGCGCATCCTTAGCGTTTCATATATGAGAATC TGGTGGTTACCCTATAGTCTGTCTGGACAGCATGCATGAAATTCATTCATGAAGGAATCA ACCAAAGCTCTCAAGCGAC
Protein Sequence (840 aa)
>BART1_0-u30899.004 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
