- Transcript BART1_0-u30899.003
- Transcript IDBART1_0-u30899.003
- Gene IDBART1_0-u30899
Exon Structure of BART1_0-u30899.003
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 593062202 | 593062067 | + |
| chr4H | 2 | 593068884 | 593066220 | + |
| chr4H | 3 | 593069956 | 593069549 | + |
| chr4H | 4 | 593070519 | 593070074 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials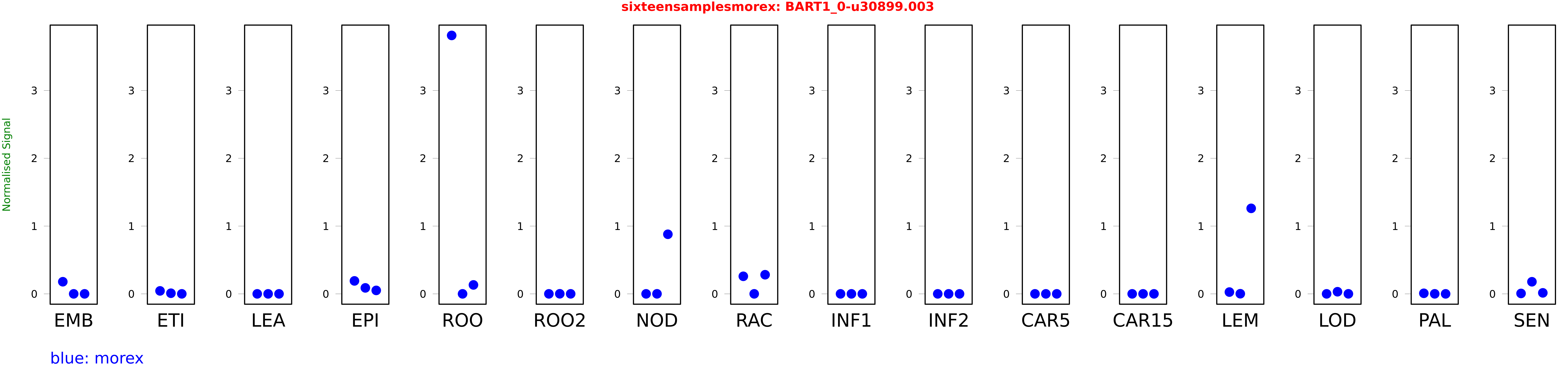
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.178773 | 0 | 0 |
| morex | ETI | 0.0437409 | 0.00924599 | 0.0000751405 |
| morex | LEA | 0 | 0 | 0 |
| morex | EPI | 0.191102 | 0.0880471 | 0.0509858 |
| morex | ROO | 3.81252 | 0 | 0.131675 |
| morex | ROO2 | 0 | 0.000983521 | 0.00023246 |
| morex | NOD | 0.000000000355588 | 0.000396357 | 0.879089 |
| morex | RAC | 0.259044 | 0.000136783 | 0.282117 |
| morex | INF1 | 0 | 0 | 0 |
| morex | INF2 | 0 | 0 | 0 |
| morex | CAR5 | 0 | 0 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 0.0263752 | 0.00179613 | 1.26148 |
| morex | LOD | 0.00000000298936 | 0.031302 | 0.000426287 |
| morex | PAL | 0.00827062 | 0.000413043 | 0.00000122869 |
| morex | SEN | 0.00611658 | 0.178408 | 0.0147356 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g38900.1 | +3 | 0.0 | 1229 | 648/853 (76%) | protein|histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH1, putative, expressed |
| TAIR PP10 | AT1G73100 @ ARAPORT AT1G73100.1 @ TAIR |
+3 | 0.0 | 556 | 272/511 (53%) | Symbols: SUVH3, SDG19 | SU(VAR)3-9 homolog 3 | chr1:27491970-27493979 FORWARD LENGTH=669 |
| BRACH PP3 | Bradi1g53840.1.p | +3 | 0.0 | 1292 | 687/855 (80%) | pacid=32801893 transcript=Bradi1g53840.1 locus=Bradi1g53840 ID=Bradi1g53840.1.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3655 bp)
>BART1_0-u30899.003 3655 150831_barley_pseudomolecules NNCCCCCCCCCCCCCCCCTGCCGTGGCGGCGCGGCGGCGCGATTCCTTTGCTGCTCGGCG GCAGCGGCCGACGGAAGCAGCGGCCGGGGTAGCAGCGACGGCAAGGCTCGGTTCTCTCCT CTCCCTCCCAACCAAGCTTTCTCCGAAGTATGGCTTGTGGACAAGTGTAGACCTGTAGAC AAAGAGACAAAGAAATTGCTGACATAAGTCCTTATTTGAGAAAGATCAGTTGTCAGCTTA CAATGGCTGGAAATCAGCAGCCGGCTTCAGTTGTGTTGGATTATGCAGCAGTCCTCGACG CCAAACCCTTGCGCACACTGACCCCCATGTTCCCTGCACCGCTAGGGATGCACACTTTCA CCCCTAAAAGCTCATCTTCAGTTGTCTGTGTCACCCCGTTTGGACCATATGCTGGAGGTA CCGAACTGGGAATGCCTGGTGGTGTTCCGCCAATGTTAACATCACCGTCCGCTTCTGCAG ATCCCAAGCAGGTGCAGCCATATACGGTTCATATGAATGGAACTTCTAATGCTAATGGTA CTACAAGCAACACAATGGTTACCCCTGTGTTGCAAACTCCTCCAGCAGTCACTACACAGG AGTCTGGTAAGAAGAAGAGGGGGATGGTTACCCCTGTGTTGCAAACTCCTCCAACAGTTA CTACACAGGAGTCTGGTAAGAAGAAGAGGGGGAGGCCCAGGCGTGTGCAAGATACAACTA CTGTTCCCCCGGTTCCTCCAGTTCAACCGGTTCCTACAGTTCATTCAGTTCCTTCAGCTC CTCCAGAAGTTAATAATATTGTTCTCCAGACACCTACTTCAGCCGTCTCACAGGAATCTG GTAAGAGGAAGAGGGGACGACCCAAGCGTGTGCCAGATGTTTCTGTCTTATCAACTCCAG TGCCTGTAGCAGATGGTACGCCTATTTTACAGACACCTCCTGCATCTAGTGTACATGAAT CTGTTACAAAGAAAAGGGGGCGGCGATCCAAGCTTGTGCAGGATATTTCAGATACTTCAA CTCCCCCAGTTCATTCAAAAGAGAGTGAGCCCTTTATGCAGACTCCTGCAGTCACCGTAT TGGAGGATGGTAAGAGGAAGAGAGGGCGGCCGAAGCGTGTGCCTGATAGTTCAGTGACTC CTTCAAGTCATTCTGTCCTTACAGTAGATGTTGACAGTGGTGACACATCAAAACGTGGCC GGCCTAGAAAAATTGACACAAGCCTATTGCACCTGCCATCTTTGTTTTCAGACGATCCCA GGGAATCCACAGATAATATACTTATGATGTTTGATGCACTGCGACGGCGGCTCATGCAGC TGGATGAGGTGAAGCAAGTAGCAAAACAGCAGCAAAACTTGAAGGCTGGGAGTATCATGA TGAGCGCTGAACTTCGCCTCAGTAAGAACAAGAGGATTGGAGAGGTTCCAGGTGTTGAGG TTGGTGACATGTTCTACTTCAGAATTGAGATGTGCCTGGTAGGGTTAAATAGTCAGAGCA TGGCAGGGATAGATTATATGTCTGCTAAGTTTGGTAACGAGGAGGACCCCGTGGCCATTA GTATTGTGTCAGCTGGTGTGTATGAGGATGCTGAAGATAATGATCCAGATGTTCTGGTTT ACAGTGGACATGGCATGTCTGGTAAGGATGACCAAAAGCTTGAGAGAGGTAATCTTGCAC TGGAGAGAAGTTTGCATAGGGGTAATCCCATTAGAGTCGTTCGCACTGTGAAAGACTTGA CTTGTTCAACTGGTAAGATATACATATATGATGGCCTTTACAGGATCAGAGAAGCCTGGG TAGAGAAAGGTAAATCTGGTTTCAATATGTTTAAACACAAGTTGCTCAGAGAACCTGGAC AACCTGATGGCATTGCAGTGTGGAAGAAGACTGAAAAATGGAGGGAAAATCCATCCTCTA GAGATCGTGTTATAGTGCACGACATATCATACGGTGTGGAAAGTAAGCCTATCTGCCTTG TAAATGAGGTTGACGATGAGAAAGGTCCTAGCCACTTCACGTATACAACTAAACTTAACT ACATGAATTCCCCAAGCTCAATGAGAAAGATGCAAGGCTGCAAATGCACAAGCGTATGCC TACCCGGTGATAACAACTGTTCTTGTACGCATCGAAATGCTGGTGACCTTCCTTACAGTG CTTCGGGCATACTTGTTAGTCGGATGCCTATGTTATATGAGTGCAATGATTCATGCACTT GTTTACATAATTGCCGGAACCGGGTTGTACAAAAAGGTATCAAGATCCACTTTGAGGTGT TTAAAACGGGAGATCGAGGCTGGGGCCTGCGTAGTTGGGATCCAATCCGAGCAGGCACAT TCATCTGTGAATATGCAGGTGTAATTGTTGACAAGAATGCTCTTGATGCAGAAGATGATT ATATCTTTGAGACCCCTCCTTCGGAGCAGAACTTAAGATGGAACTATGCACCAGAATTAC TGGGTGAACCTAGTCTTTCTGACTTAAATGAATCATCTAAGCAGTTGCCAATAATTATTA GTGCAAAATATACTGGTAACATAGCTCGCTTCATGAACCACAGCTGCTCACCTAATGTTT TTTGGCAACCAGTTCTGTATGACCATGGTGATGAGGGATATCCACACATCGCATTCTTTG CAATTAAGCATATTCCTCCAATGACAGAGCTTACATATGATTATGGTCAGAGCGGTAATT CAGGGTGTCGGAGGTCTAAAAGTTGCTTATGTTGGTCTCGCAAGTGCAGGGGTTCCTTTG GCTAAATAATCAACCCTTAAGGGATACTTTCCTAGTTGAAGAAGCAGCTTATGGTGGCGG CGGCATTATGGGTGTGGTGTTCCAGCCACAGTTGAAATAACTTTGCTGCCTCTTTCTATC CGGGGTTGGAATGCGGGGTAGAGTACAAACGACTAGATGTTGTAATAAAGGGCAAGCTAT GCTTCTGTGGTGTTGTTAGTTGTAGTTGTCTGCTCCGGTTTAAGACCACCCAGTGGAATT CCTTCTATGTATTGTCATTGCGAAGATGTTTCAAGCCTGCAGTGAATCTATCGTTTTCGA TTTGTAATCCTAGCGGATTAATGTGTCCTGTGAATCTGTGATTGCCGTGGAGAGCGAATG AAGGCAACAATAATAATCCAGCTGGGCATTGTTCGTGCAGGGGTAAAGATAGGGGCAGAG TCAGTGCTGTGCGCTTGATTTATGATTCGATTTGGTGTGAAGGATGGTGCTCTGTTCCTA TGAGCAGTAGGCTCGCTGGGATTTTGGGCTTTGCCGTGTATTTTGGTCCCTGCTTGGCAT ACCGTTTTCTGCACATTACAGGTGTAAAATAAATACAGACCTCCATCCACTGGTTTAGTT TCCAGCCGGAAGACACTCATGTCCAGTAAAATACACCTGTAAATTTTACATCCCTGTATT AAATTACACCTATACCAAGCGCATCCTTAGCGTTTCATATATGAGAATCTGGTGGTTACC CTATAGTCTGTCTGGACAGCATGCATGAAATTCATTCATGAAGGAATCAACCAAAGCTCT CAAGCGACGCATATGTTCATCTAGCATTTCATATCCGAACTTGATATGATACATGAGAAT AAGGCCCAATTCTCTCAGAATCTAGGGATTCTCTCATAATCAAATCCCTTCCCAG
Protein Sequence (840 aa)
>BART1_0-u30899.003 840 150831_barley_pseudomolecules MAGNQQPASVVLDYAAVLDAKPLRTLTPMFPAPLGMHTFTPKSSSSVVCVTPFGPYAGGT ELGMPGGVPPMLTSPSASADPKQVQPYTVHMNGTSNANGTTSNTMVTPVLQTPPAVTTQE SGKKKRGMVTPVLQTPPTVTTQESGKKKRGRPRRVQDTTTVPPVPPVQPVPTVHSVPSAP PEVNNIVLQTPTSAVSQESGKRKRGRPKRVPDVSVLSTPVPVADGTPILQTPPASSVHES VTKKRGRRSKLVQDISDTSTPPVHSKESEPFMQTPAVTVLEDGKRKRGRPKRVPDSSVTP SSHSVLTVDVDSGDTSKRGRPRKIDTSLLHLPSLFSDDPRESTDNILMMFDALRRRLMQL DEVKQVAKQQQNLKAGSIMMSAELRLSKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSM AGIDYMSAKFGNEEDPVAISIVSAGVYEDAEDNDPDVLVYSGHGMSGKDDQKLERGNLAL ERSLHRGNPIRVVRTVKDLTCSTGKIYIYDGLYRIREAWVEKGKSGFNMFKHKLLREPGQ PDGIAVWKKTEKWRENPSSRDRVIVHDISYGVESKPICLVNEVDDEKGPSHFTYTTKLNY MNSPSSMRKMQGCKCTSVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSCTC LHNCRNRVVQKGIKIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGVIVDKNALDAEDDY IFETPPSEQNLRWNYAPELLGEPSLSDLNESSKQLPIIISAKYTGNIARFMNHSCSPNVF WQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSGNSGCRRSKSCLCWSRKCRGSFG
