- Transcript BART1_0-u30598.009
- Transcript IDBART1_0-u30598.009
- Gene IDBART1_0-u30598
Exon Structure of BART1_0-u30598.009
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 574820351 | 574820286 | + |
| chr4H | 2 | 574820541 | 574820449 | + |
| chr4H | 3 | 574820930 | 574820755 | + |
| chr4H | 4 | 574821065 | 574821019 | + |
| chr4H | 5 | 574822272 | 574822203 | + |
| chr4H | 6 | 574822810 | 574822589 | + |
| chr4H | 7 | 574822997 | 574822908 | + |
| chr4H | 8 | 574823189 | 574823114 | + |
| chr4H | 9 | 574823585 | 574823332 | + |
| chr4H | 10 | 574829176 | 574828831 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials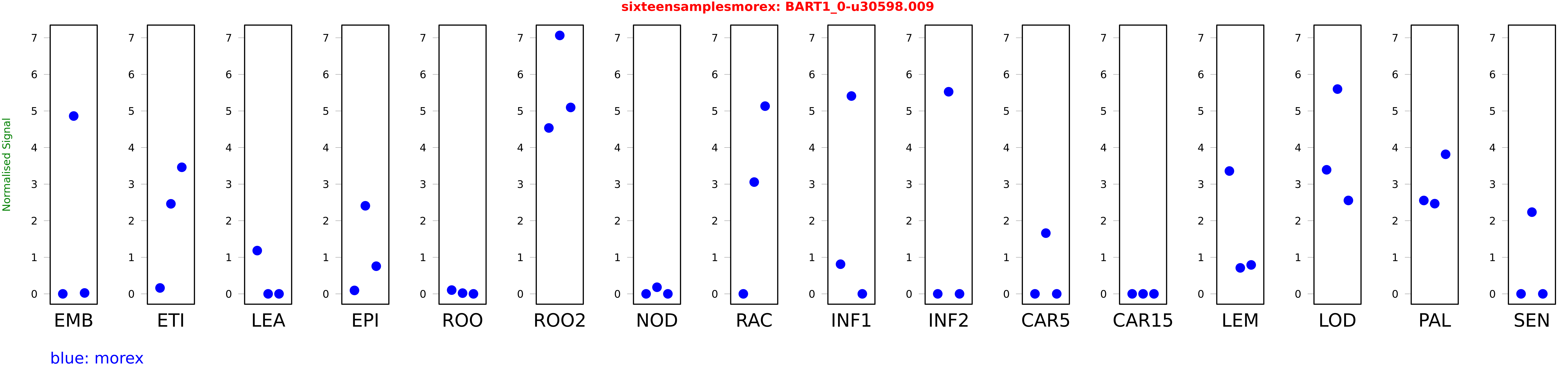
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.0000000299839 | 4.86108 | 0.0243273 |
| morex | ETI | 0.161402 | 2.46094 | 3.4601 |
| morex | LEA | 1.18263 | 0 | 0 |
| morex | EPI | 0.0941727 | 2.40748 | 0.756253 |
| morex | ROO | 0.102903 | 0.0197929 | 0.000000405654 |
| morex | ROO2 | 4.53493 | 7.0645 | 5.09607 |
| morex | NOD | 0 | 0.180849 | 0 |
| morex | RAC | 0.000000220563 | 3.05539 | 5.1317 |
| morex | INF1 | 0.81078 | 5.40761 | 0 |
| morex | INF2 | 0 | 5.52489 | 0 |
| morex | CAR5 | 0.00000000493144 | 1.65995 | 0 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 3.35783 | 0.710134 | 0.791038 |
| morex | LOD | 3.39114 | 5.59797 | 2.55356 |
| morex | PAL | 2.55278 | 2.46548 | 3.81522 |
| morex | SEN | 0 | 2.23377 | 0.000000000599911 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os03g10460.1 | +3 | 2e-128 | 286 | 142/176 (81%) | protein|expressed protein |
| TAIR PP10 | AT2G18410 @ ARAPORT AT2G18410.1 @ TAIR |
+3 | 3e-54 | 130 | 76/181 (42%) | Symbols: | FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: cellular_component unknown; EXPRESSED IN: 22 plant structures; EXPRESSED DURING: 13 growth stages; CONTAINS InterPro DOMAIN/s: Histone acetylation protein 2 (InterPro:IPR019519); Has 35333 Blast hits to 34131 proteins in 2444 species: Archae - 798; Bacteria - 22429; Metazoa - 974; Fungi - 991; Plants - 531; Viruses - 0; Other Eukaryotes - 9610 (source: NCBI BLink). | chr2:7990768-7992588 FORWARD LENGTH=374 |
| BRACH PP3 | Bradi1g71057.2.p | +3 | 4e-144 | 299 | 147/176 (84%) | pacid=32795801 transcript=Bradi1g71057.2 locus=Bradi1g71057 ID=Bradi1g71057.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (1440 bp)
>BART1_0-u30598.009 1440 150831_barley_pseudomolecules GCGCGCCATGCTCCACGTCGCCGCCGCCGTCGCCTCCAACGCGGCCGCGGGGAAGGCGCA GGCCAGGGGACTTGTGGTTGTGGCGTTTGACCGGAGCCCGGAAGTTTACCTGGATGCCAT GCTTCGGCACGGCCTTGATTCGAATGCACTGAGTCGATGTGTTCGGATATTGGATTGCTA CTCAGACCCACTCGGGTGGAAGAAAAATATCCGAAGTCAGCAGCATCATGAGGACAGTGG AACACCGTGCTCATCAAACAAAGGCAACATCACAATTTTCAGAAGCGTGAAGGATGTTGA TAAGTTATCGTGCTCCATAATTGATCTTGGAAGAGGGTTTGAAGGAGAAGGCAAGACCTA TTTTTCTGTTGCTCTCGATTCAATTAGCTCTATGTTAAGGCATGCTTCAGTGCAATCAAT TTCAGGCCTTCTTAGTAATCTTCGGAGCCATGATATCATCAATCTTCTGGCTAATGCATT CAGATCTCCATGAAACCAAGTTTTCCCGAGCTTTTGAATGCCTTTCTACCATGGTTGCTT GTGTAGAGCCAGAAGTTGTAGATTCTGTATATGGAGAAGAACGTAGGGGGGACATGTCTT TCCTTGAGCATAACTATTCGAAAGCAAAGTTCCATGTGCGCCTGAAACGAAGGAATGGAC GGGTGAAGCATCTGCATGAGGAGTTATGTGTTGAGGGATATGATGTAAAATTTGTTTCTG CTACTTCTGTAAGTATGGAAGTGAACCAAAGTCTACTACCTAAGGTTCAGTTCAATCTTG AGCTGTCAGAAAAGGAACGGAGTGACAGGGCAAATGTTGTTCTTCCTTTCGAACATCAAG GAAAGGGCGAGCCAATCCGTATATATGATGGACGGCGATCTCTCCCGGAGGCTCGTCAAG ATCTCAATTTTACTACGGCAGCTCTCCTGGATGAAACAGAAGCTCTTAAATCTGCAGACA CGAAGGGTGAAATTCATTACCTTCGTGATTCTGATGATGAGCGACCTGACTCGGATGAAG ACCCCGATGATGATTTGGATATTTAGCCACAATGTGAGCTCTGTTTATCCTAGTGGTCAG CTATCAAGTCAAAGGAACATCAGGCGCTTGCGAAAATTCTAGGAGGCGTTCACTTTGCGT GGGCGGCTGGAGCTACTGCATAGTTATGGTTGAACAATTTAAGCACCTGTATTATGTATA GCTTCGTGAGCATTGGAAGGCAACCTTTGCAAATCACTGGGTTTGAAATTTCTTGGATGC TCAAGACTAATGTGTTGATGGTAGCATATCGAATGAAAGGTCATATGCCCCAATGCGATT TTGTAGTTTTGTTTCGACGTGGATGAATGTTGGCTAGTGTGTCAGTCGCTGAAATGGGTC CAGAACAAGAGGATGCGACATGTAACACTGTTTTTTCAGTTATATGAGAATATACACCGG
Protein Sequence (214 aa)
>BART1_0-u30598.009 214 150831_barley_pseudomolecules MLQCNQFQAFLVIFGAMISSIFWLMHSDLHETKFSRAFECLSTMVACVEPEVVDSVYGEE RRGDMSFLEHNYSKAKFHVRLKRRNGRVKHLHEELCVEGYDVKFVSATSVSMEVNQSLLP KVQFNLELSEKERSDRANVVLPFEHQGKGEPIRIYDGRRSLPEARQDLNFTTAALLDETE ALKSADTKGEIHYLRDSDDERPDSDEDPDDDLDI
