- Transcript BART1_0-u27957.023
- Transcript IDBART1_0-u27957.023
- Gene IDBART1_0-u27957
Exon Structure of BART1_0-u27957.023
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 166877905 | 166877543 | - |
| chr4H | 2 | 166878371 | 166878131 | - |
| chr4H | 3 | 166884880 | 166881947 | - |
| chr4H | 4 | 166885393 | 166884994 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials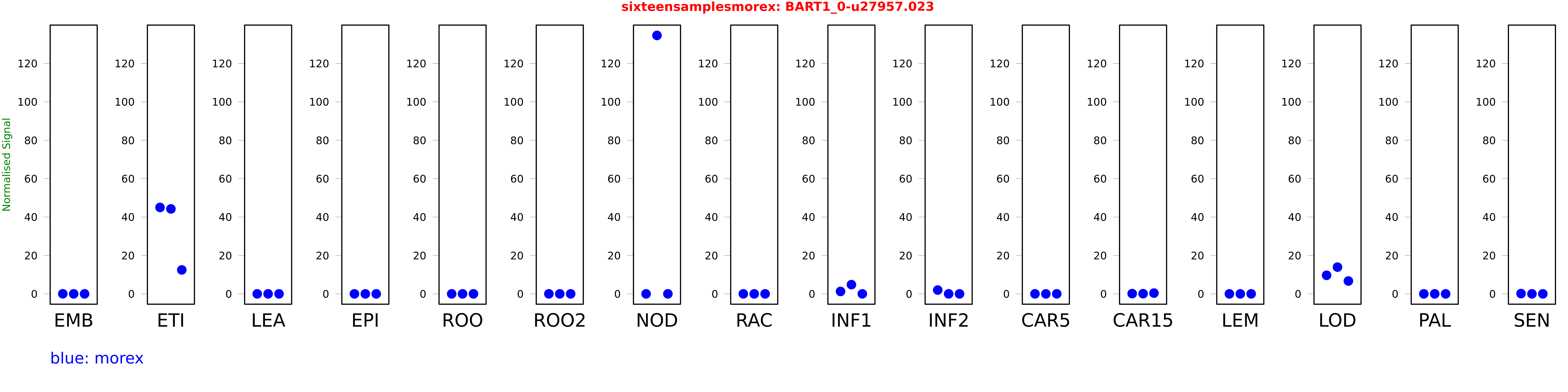
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0.0351508 | 0 | 0 |
| morex | ETI | 45.0174 | 44.2396 | 12.4738 |
| morex | LEA | 0.000800668 | 0.000000157762 | 0 |
| morex | EPI | 0 | 0 | 0 |
| morex | ROO | 0 | 0 | 0.00833143 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 134.603 | 0 |
| morex | RAC | 0 | 0 | 0 |
| morex | INF1 | 1.25745 | 4.80218 | 0.00841271 |
| morex | INF2 | 1.97884 | 0.00128192 | 0 |
| morex | CAR5 | 0.00000000936734 | 0 | 0.0167507 |
| morex | CAR15 | 0.120848 | 0.0930219 | 0.391685 |
| morex | LEM | 0 | 0 | 0 |
| morex | LOD | 9.67046 | 13.9209 | 6.68806 |
| morex | PAL | 0 | 0 | 0 |
| morex | SEN | 0.13798 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g47970.1 | +2 | 0.0 | 751 | 360/420 (86%) | protein|AAA-type ATPase family protein, putative, expressed |
| TAIR PP10 | AT2G39730 @ ARAPORT AT2G39730.1 @ TAIR |
+2 | 0.0 | 758 | 365/468 (78%) | Symbols: RCA | rubisco activase | chr2:16570951-16573345 REVERSE LENGTH=474 |
| BRACH PP3 | Bradi4g09125.2.p | +2 | 0.0 | 844 | 404/457 (88%) | pacid=32783473 transcript=Bradi4g09125.2 locus=Bradi4g09125 ID=Bradi4g09125.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3938 bp)
>BART1_0-u27957.023 3938 150831_barley_pseudomolecules CGCAGGGACGCTCTAGAATAGTCTCTCACGCTCTAGGTCACTCCAGTCGTGTTGGACGCT TGCTCCATGGCCATCTGTCCATGCACCTATATAAGACGCCATGGCCAAGCTCCATCGATC GGCATCGCATCTGTACCTGCTAACCACTGCACCCCTCTCTTTTATCCATATCAGCTCCAT CCATCACTGACGATCGATCTCAGTCCACCTGCATCTGTAGGTGTACTAGTAGTACGCCGG CCTAATCCAGCTGAGAGCTGATCGAGCTAGCTTGCATATACCACTGCACGTACGCACATC AAATTAATTAGAAACTACTGTAATTAAGTTATCAGCAGCGAGAAGAGAACGATCGATCAA CACCATGGCTTCTGCTTTCTCGTCCACCGTTGGAGCTCCGAGTTGCTACACATTTTCAGG TCTGAATTGTTTCATTTTGTACAGTTTTTAATATAATTTGTGACCGTGTGATCTTGCTTT GAGATTCGGACGCGACAAGCTGGGCCTCCTCTCTATGTGTTGACTCCGACAAGGGCATCT CTTTCCTCACCCATGCCTTCCGGGGCTTAAAAAGACTGTCTATGGTCAGCCTGAGCGTTC GGGTATTTTGGTTATTTCGGTTCATGTGTAATTCGGGTATTAAAAAAATGGGAATCGGAA TTTTTTATGTCAAAAAAATAACCGAACCCAAATTACCCAAAATTTTGGTTTGGGTAACTC GGGTACCCAAAAAACCCAAAATATTCAGAGCACACATTAAGTTTCGGCATGAATAATTCA GATAGTGTGGGTGTTTTAGTTAGTTTGTGTAACATGTAAGTGTAATAATAACATAAATAA TTTGACTAGTATGAATAATTCGGGTAGTATGGGTATTTTGGGTTTTACAGACTAGAACCT GAACCCTAACCCGAAAACCGAATAGCCAGGAAATGAGAACCAAACCTTAACCCGATACCC ATATTACCCGACAATTCGGGTATTTTGGTTCAGTTCGGGTTTGGGTAGCGGGTTAGGGTA TCCAAACTCTCGGGTGAGTCTAGGGTGATGGGGTGGGCAGCGGATGGCTAGCCACCATGG GGCGAAGGCTATCAAATGAGGTCATTTGATCTCGGTTTGGGCATGACCTTGAATATTGAG TTAGCACGATAAAGATAATTTAGGCAATACTAATGGTTGTTTCATGATGGCGCGGATGAG AATGAACGGGAGGTGGTAGCGAGATTTTATTTAGGAGCAACTATTGTGAGGTTTATTAAA TCACGATGCAAATTACAAATAGGGCGAGGTGAGGGTGGTGACGGGGAGTGTTGAGAAATG TGGGAAGCTAATGGTTACACTTTTTCGAAGCTATTGCGTGGAGGAGGAAGGGAAAGATAG AACCTCAAAAGTCTCGAGTGGAGGCACACGGTTGTCAAGCCCGTGTGTGAGAATNATAAT ATTAGAAACTAACTAGTGAAGTATGCTTCTCCTCATTCTTCATTAGTAAGAACTTTTGGC TGAATTAGTTACTCCCTCCGTCTCAGTGTATAGGTCATATTAGGTTGTGCATCGTGATCA AAACGGAGAGGAAAACAAAAAAATATATTAATTTGCTAATTAATATCATTGCATGTAATG AATTAACCACTGCATGTCGTGCTAGTTAGTCGCAAGTCATTAAAAACATACACGTTCCAT GTCTCTTATTGGTTGATTCCTTTATCCATGTAAAAAAAAACAAAAAACAAGCAGAAAATT AATGCATCGTGCCTAAGTGTTTTGAGATTTTTTTTTCATAAGATGACTTATAAACCAAGA CGGAGGGAGTAGGTTGTTTTGTAATTCAATATGTCATTCAAGCCATGGGGCATCAACTTC AAGGCCCAGCATGCCGGGACGGGACGGATCTAAAAACTATTTTGACTCGTGGCAAAACAT CATTAAAATAAGGGAGGTAAATGATGGAGAGTAACAAAAACGATTTAAGAGTGGTGTCAC ATTTTATCCAAATTCATTGAGTTTTAGCATTTCCTCCTAAAAGTTATTCAAATCGTATTG AAATTTCATATTTTGAGTAGGGTCCTAGGCCCCCACTCTCTTCAACACGGGCCCACCACT TCTGGTATTGTACCTTTGACCTTCCGCCCCTGTTCATCAGCTGGAGCTTTTGGATGGACT AATTAATTCTATATATATGCGGATGCAATTGAATAGCGAGTGAAGTCTGGAACTGCCCAA ATGAAGGGTTGATATCTGCAAATTAAGCGTTCATCGTGGCAATTGCTATTGAAAGCAAGC AGCAAGTGAACCAACGTGCATGAATGTGATTGGGGCTGACTATGTCATACTTGTGCAGGC GTCGACCCCGACCATCTTCCTGGGCAAGAAGGTGAAGAACTACTACCATGGTGGCAACAA GATGAAGAGCAGGGTGGTGAGGGTCATGGCGGCCAAAAAGGAACTTGACCAGGGCAAGCA GACCGATGCGGACCGGTGGAAGGGTCTGGCCTACGACATCTCGGACGACCAGCAGGACAT CACTCGGGGGAAAGGCATCGTGGACTCCCTGTTCCAGGCCCCCATGGGCGACGGCACCCA CGAGGCCATCCTAAGCTCCTACGAGTACATCAGCCAGGGCCTGCGGAAGTACGACTTCGA CAACACCATGGACGGGCTGTACATCGCGCCGGCATTCATGGACAAGCTCATCGTCCACCT CGCCAAGAACTTCATGACACTCCCCAACATCAAGGTTCCTCTCATCCTGGGTATCTGGGG AGGCAAGGGACAGGGCAAGTCGTTCCAGTGTGAGCTGGTGTTCGCCAAGATGGGCATCAA CCCCATCATGATGAGCGCCGGTGAGCTGGAGAGCGGCAACGCCGGCGAGCCGGCCAAGCT GATCCGGCAGAGGTACCGCGAGGCGGCCGACATTATCAACAAGGGCAAGATGTGCTGCCT CTTCATCAACGACCTGGACGCCGGCGCGGGCCGGATGGGCGGGACGACGCAGTACACGGT GAACAACCAGATGGTGAACGCCACCCTGATGAACATCGCGGACGCGCCCACCAACGTGCA GCTCCCGGGGATGTACAACAAGGAGGAGAACCCCCGCGTGCCCATCATCGTCACCGGCAA CGACTTCTCGACGCTGTACGCTCCCCTGATCCGTGACGGGCGCATGGAGAAGTTCTACTG GGCGCCCACCCGCGAGGACCGCATCGGCGTGTGCAAGGGCATCTTCCGCACCGACAACGT CCCGGACGAGGCCGTGGTGAGGCTGGTGGACACCTTCCCGGGGCAGTCCATCGACTTCTT CGGCGCGCTGCGGGCACGGGTGTACGACGACGAGGTGCGCAAGTGGGTCGGCTCTACCGG AATCGAGAACATTGGCAAGAGGCTGGTGAACTCGCGGGACGGGCCCGTGACCTTCGAGCA GCCAAAGATGACAGTCGAGAAGCTGCTAGAGTACGGGCACATGCTCGTCCAGGAGCAGGA CAATGTCAAGCGTGTGCAGCTTGCTGACACCTACATGAGCCAGGCAGCTCTGGGTGATGC TAACCAGGATGCGATGAAGACTGGTTCCTTCTACGGTAAAGGAGCACAGCAAGGTACTTT GCCCGTGCCGGAAGGTTGCACCGACCAAAATGCCAAGAACTACGACCCAACGGCAAGGAG CGACGACGGCAGCTGCCTTTACACCTTTTAAGCATGCCACTTTTAATATCTGATGTTTTA CCTACCTAAGTCCGCATCGCATCCAAGCGATTGCTGGGCATGTTTTGTTAATATATCTCT AAATATATTGAGTATACCCATTTGGCAAAGACCAATACACCTACTTATCATTTGTCACTC TCAGTATTCATTCCTTCAGTGGAGACTCCTTCTGCTTTGTTGCAAACAGAACAAGAAAGT ATTAATTTGAGCTTCAGCTTATTAATTATTATTGCACA
Protein Sequence (463 aa)
>BART1_0-u27957.023 463 150831_barley_pseudomolecules MNVIGADYVILVQASTPTIFLGKKVKNYYHGGNKMKSRVVRVMAAKKELDQGKQTDADRW KGLAYDISDDQQDITRGKGIVDSLFQAPMGDGTHEAILSSYEYISQGLRKYDFDNTMDGL YIAPAFMDKLIVHLAKNFMTLPNIKVPLILGIWGGKGQGKSFQCELVFAKMGINPIMMSA GELESGNAGEPAKLIRQRYREAADIINKGKMCCLFINDLDAGAGRMGGTTQYTVNNQMVN ATLMNIADAPTNVQLPGMYNKEENPRVPIIVTGNDFSTLYAPLIRDGRMEKFYWAPTRED RIGVCKGIFRTDNVPDEAVVRLVDTFPGQSIDFFGALRARVYDDEVRKWVGSTGIENIGK RLVNSRDGPVTFEQPKMTVEKLLEYGHMLVQEQDNVKRVQLADTYMSQAALGDANQDAMK TGSFYGKGAQQGTLPVPEGCTDQNAKNYDPTARSDDGSCLYTF
