- Transcript BART1_0-u27304.003
- Transcript IDBART1_0-u27304.003
- Gene IDBART1_0-u27304
Exon Structure of BART1_0-u27304.003
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr4H | 1 | 75934498 | 75931461 | - |
| chr4H | 2 | 75934901 | 75934770 | - |
| chr4H | 3 | 75936428 | 75936189 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials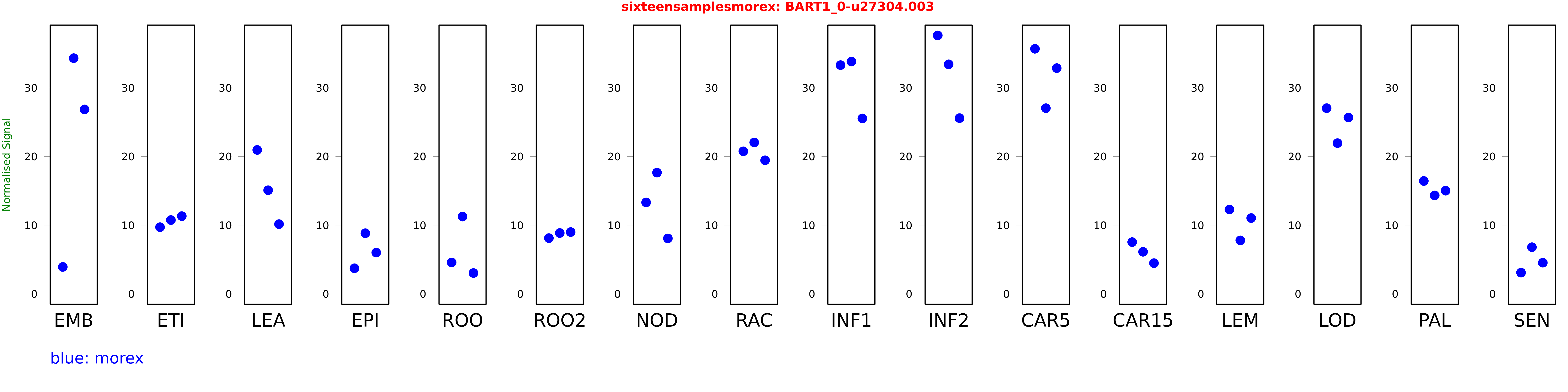
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 3.92119 | 34.3392 | 26.8766 |
| morex | ETI | 9.71528 | 10.7534 | 11.3246 |
| morex | LEA | 20.965 | 15.0985 | 10.158 |
| morex | EPI | 3.73129 | 8.83445 | 6.01139 |
| morex | ROO | 4.57762 | 11.2552 | 3.0349 |
| morex | ROO2 | 8.12042 | 8.86452 | 9.00117 |
| morex | NOD | 13.3186 | 17.6659 | 8.07759 |
| morex | RAC | 20.7704 | 22.0565 | 19.4589 |
| morex | INF1 | 33.3231 | 33.8405 | 25.5614 |
| morex | INF2 | 37.6509 | 33.4442 | 25.5999 |
| morex | CAR5 | 35.7048 | 27.0497 | 32.8858 |
| morex | CAR15 | 7.53825 | 6.12805 | 4.47515 |
| morex | LEM | 12.2902 | 7.79627 | 11.0421 |
| morex | LOD | 27.052 | 21.9597 | 25.6894 |
| morex | PAL | 16.4427 | 14.3406 | 15.0268 |
| morex | SEN | 3.09321 | 6.79826 | 4.53806 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os11g08090.1 | +3 | 0.0 | 1524 | 845/1007 (84%) | protein|expressed protein |
| TAIR PP10 | AT1G77460 @ ARAPORT AT1G77460.2 @ TAIR |
+3 | 0.0 | 950 | 565/1012 (56%) | Symbols: | Armadillo/beta-catenin-like repeat ; C2 calcium/lipid-binding domain (CaLB) protein | chr1:29104378-29111580 FORWARD LENGTH=2136 |
| BRACH PP3 | Bradi4g23680.2.p | +3 | 0.0 | 1570 | 891/1012 (88%) | pacid=32790009 transcript=Bradi4g23680.2 locus=Bradi4g23680 ID=Bradi4g23680.2.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3410 bp)
>BART1_0-u27304.003 3410 150831_barley_pseudomolecules GTCCATGTCCGCTGCCGTAATAACGCGACAATGACGGGAGTGATCCGCAAAAGACCCACC CCTCTCTCTCCCTCCCTCCCCACTTCCCCTGTTCCAAAAAGCTTTCGCATCTCTCTCTCT CTCTCTTCACAGTTCACACCCACACTGCGCGGCACAGTGGGCGAGTGGGCGCGAGACCAA GCGTCGCCTCCTCCTCTCCTCCGCCTCGGTCTCGCCGCCGGACGTCTTCGCCTGCCGCCG GAGGACATATAAAACAATTAAGATTGTAAACGTTGGTAACTGTTCAATCATGCCGATATC CAACTCAACGGAACCTCGAGATTCTCGAGACCCTACTTCACCTGCGCCATCCACGTCATC TTCTAGATCAAGAAGTTGATGACCCTGAAAGTGCAATGTCCACTGTTGCTCGATTGCTGG AGCAGCTACATGGTAGCTTGACATCACTGCCAGAGAAGGAAGTGGCAATCAAAAGATTAC TTGAACTTGCCAAGGCGAAGAAGGATGCAAGGATTTTGATAGGTAGCCATTCTCAGGCCA TGCCATTATTTATATCTATCCTTAGAAGCGGGGCATCTTCAGCCAAAGTAAATGCTGCGG CCCTGCTTAGTGCACTCTGCAAGGAAGAGGATCTGCGTGTCAAGGTTCTCTTAGGAGGTT GCATACCACCCTTGCTCTCTCTCTTGAAGTCTGAATCTGCTGAAGCAAAGAAGGCTGCCG CTGAGGCTATTTTTGAAGTGTCTTCAGGGGGTCTTTCGGATGATCACATCGGCATGAAAA TATTTGTGACAGAAGGCGTTGTGCCGACTCTTTGGGATTTGCTCAATCCTAAGTCACGCC AAGATAGAATAGTTGAGGGCTTTGTGACTGGGGCTTTAAGAAATCTCTGTGGAGATAAAG ATGGTTATTGGAAGGCCACACTTGAAGCGGGTGGAGTAGAAATTATTACCGGTCTTCTTT CATCCAAAAATACTGCTTCGCAGTCAAATGCTGCCTCCCTCTTGGCACGGTTGATTTCTG CTTTCAGTGATAGCATTCCCAAAATCATAGCTGCTGGGGCTGTTAAGGCCCTTCTTCAGC TTCTGAATCGAGATGATGACATTGCTGTTCGTGAAAGTGCAGCTGATGCTTTGGAGGCCT TATCTTCCAAGTCTACCATTGCAAAGAAAGCTGTGGTCGATGCAGGCGGCCTCCCTGTTT TGATTGGAGCTGTTGTAGCGCCCTCAAAAGAGTGCATGCGAGGGGTTACCTGCCATTCTC TACAAAGCCATGCCGTTTGTGCTTTATCAAATATTTGCGGTGGAGCCACTTCGTTGTTGC TTTACCTGGGAGAGCTTTGCCAATCACCCCGGTCAGCTGTGTCCCTTGCTGACATTCTTG GAGCACTTGCATATACGCTGATGGTGTATGATGGCACTGATGGTAAATTCTTTGATCCTG TTGAGATAGAAAGCATTTTGGTTGTGCTCCTGAAATCCCATGACAGTAAGCTTTTGCTTG ATCGTATTCTTGAAGCTTTAGCAAGTCTATATGCAAATGCATGTTTCTCTGGCAGACTTG ATCACTCAAATGCAAAGAAGGTTCTTGTTGGGCTGGTCACTATGGCTACCGACGATGTTC AAGACCATCTTGTTCATGCCTTAACTAGCTTGTGCTGTGATGGTTTTGGATTGTGGGACG CTCTTGGAAAGAGAGAAGGAGTTCAGTTGCTAATATCATTACTTGGACTTTCCAGTGAGC AGCATCAGGAGTATGCAGTTTCTTTGTTGGCTATCTTGAGTGATGAAGTAGATGACAGTA AGTGGGCAATAACAGCTGCTGGAGGTATCCCTCCACTTGTCCAACTACTTGAAACAGGAT CTCAAAGGGCAAAGGAGGATGCAGCTCATATCATATGCAACTTGTGCTGTCACAGTGATG ATATCAGGGCATGTGTTGAGAGTGCTGGTGCTGTTCTGGCGCTGCTTTGGCTTCTAAAGA GTGACAGTCCTCGTGGACAAGAGGCGTCAGTCAAAGCACTCAAGGTGCTTATACGGTCTG CTGATTCCGCCACAATCAATCAGCTGTTAGCACTGCTGCTCAGTGACTCAGTGAGCTCAA AGGCACACGCCATTACAGTTCTTGGGCATGTCCTTGTGCTGGCTCCTCAAAGAGACCTAA TCCAGAATGGAGCTCCTGCTAATAAAGGGCTTAGGTCTCTTGTTCTTGTTCTTGACTCGT CAAATGAAGAATCTCAGGAATGTGCTGCAACTGTGTTGGCTGATATTTTCAGTATGCGTC AGGATATTTGCGACATCTTAGCAACTGATGAAATTGTTCAGCCATTCATGAAGCTTTTGA CAAGTGGAAATCAAGTTATGGCAACACAATCTGCTCGAGCTTTAGGAGCACTCTCGCGCT CGGCTAATACCATGTCGAAGAACAAGATGTCATGTATAGCTGAAGGTGACGTGCAACCTC TCATAGAGATGGCCAAGACATCATCCATTGATGCGGCTGAAGCGGCAATTGCTGCATTGG CAAATCTCTTATCAGATTCCCAGATTGCTAAGGAGGCGCTAGGTGATAATATTGTCCAAG CTTTGACTAGAGTCCTAAAGGAGGGGAGTTTAGATGGCAAAATTAGTGCTTCACGGTCAC TTTATCACTTGCTCAACCAATTCCCCCTCTGTGAAGTTTTTCCAGATTACTCACTCTGCT GCTTTATAATTCATGCACTGCTGGTGTGTCTATCCGGTATCAGTTTGGAAAAAGTTACTA GTTTGGATCCCCTGGATGTGCTTGCATTGATGGTGATGACCAAGGAGGGTGCTCATTTTA GCCCCCCTCTGCGGACTGCCTTTCTTGAAGCCCCAGAAGGTTTAGAACCTCTGGTCCGCT GTATCAGTGTTGGGCTCCCCCCTATTCAAGACAAATCCATTCAAATTCTCGCAAGGCTAT GTCAGGACCAATCTTCTCTGCTTAGTGAGCACATAAACAGAAGTGAAGGCTGCATTGATT CTCTTGTCAGTAGAGTTATGGAATCAACAAACATGGAAATAAGAATCTCCAGTGCAATCA CCCTCATATCTGCGTTGAAGGACAAGAGAGAACATTCAATTGAAGTTCTTGAAGCATCTG GACATCTGAAGAGTCTAATATCTGCACTCATTGATATGTTGAAGCAAGAGTCCACTTCAA CATCCCTAGATATTGAAGTCTGGAAACCTTACACAGAGAAGAGTTTGTTCAATTGTGAGC AGGATGTTTTGGATGTGCCTGAATCAGGGAAGGTTCTGGAAGAAACTGTTGCACGTTTGC TCTCACTAATTTGTTCTTCTCATCCCAGGAGTAAACTTACTGTTATGGATCTTGGTGGTG TTGAGATTGTTTCTGATAAACTTGCTTCTGCTAGCCGACAGGTGCATGTT
Protein Sequence (1005 aa)
>BART1_0-u27304.003 1005 150831_barley_pseudomolecules MSTVARLLEQLHGSLTSLPEKEVAIKRLLELAKAKKDARILIGSHSQAMPLFISILRSGA SSAKVNAAALLSALCKEEDLRVKVLLGGCIPPLLSLLKSESAEAKKAAAEAIFEVSSGGL SDDHIGMKIFVTEGVVPTLWDLLNPKSRQDRIVEGFVTGALRNLCGDKDGYWKATLEAGG VEIITGLLSSKNTASQSNAASLLARLISAFSDSIPKIIAAGAVKALLQLLNRDDDIAVRE SAADALEALSSKSTIAKKAVVDAGGLPVLIGAVVAPSKECMRGVTCHSLQSHAVCALSNI CGGATSLLLYLGELCQSPRSAVSLADILGALAYTLMVYDGTDGKFFDPVEIESILVVLLK SHDSKLLLDRILEALASLYANACFSGRLDHSNAKKVLVGLVTMATDDVQDHLVHALTSLC CDGFGLWDALGKREGVQLLISLLGLSSEQHQEYAVSLLAILSDEVDDSKWAITAAGGIPP LVQLLETGSQRAKEDAAHIICNLCCHSDDIRACVESAGAVLALLWLLKSDSPRGQEASVK ALKVLIRSADSATINQLLALLLSDSVSSKAHAITVLGHVLVLAPQRDLIQNGAPANKGLR SLVLVLDSSNEESQECAATVLADIFSMRQDICDILATDEIVQPFMKLLTSGNQVMATQSA RALGALSRSANTMSKNKMSCIAEGDVQPLIEMAKTSSIDAAEAAIAALANLLSDSQIAKE ALGDNIVQALTRVLKEGSLDGKISASRSLYHLLNQFPLCEVFPDYSLCCFIIHALLVCLS GISLEKVTSLDPLDVLALMVMTKEGAHFSPPLRTAFLEAPEGLEPLVRCISVGLPPIQDK SIQILARLCQDQSSLLSEHINRSEGCIDSLVSRVMESTNMEIRISSAITLISALKDKREH SIEVLEASGHLKSLISALIDMLKQESTSTSLDIEVWKPYTEKSLFNCEQDVLDVPESGKV LEETVARLLSLICSSHPRSKLTVMDLGGVEIVSDKLASASRQVHV
