- Transcript BART1_0-u25435.016
- Transcript IDBART1_0-u25435.016
- Gene IDBART1_0-u25435
Exon Structure of BART1_0-u25435.016
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr3H | 1 | 681728097 | 681727681 | - |
| chr3H | 2 | 681728628 | 681728591 | - |
| chr3H | 3 | 681728830 | 681728758 | - |
| chr3H | 4 | 681729054 | 681728926 | - |
| chr3H | 5 | 681729370 | 681729308 | - |
| chr3H | 6 | 681729697 | 681729472 | - |
| chr3H | 7 | 681730030 | 681729865 | - |
| chr3H | 8 | 681730185 | 681730105 | - |
| chr3H | 9 | 681730317 | 681730271 | - |
| chr3H | 10 | 681730783 | 681730695 | - |
| chr3H | 11 | 681731206 | 681731096 | - |
| chr3H | 12 | 681732647 | 681732395 | - |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials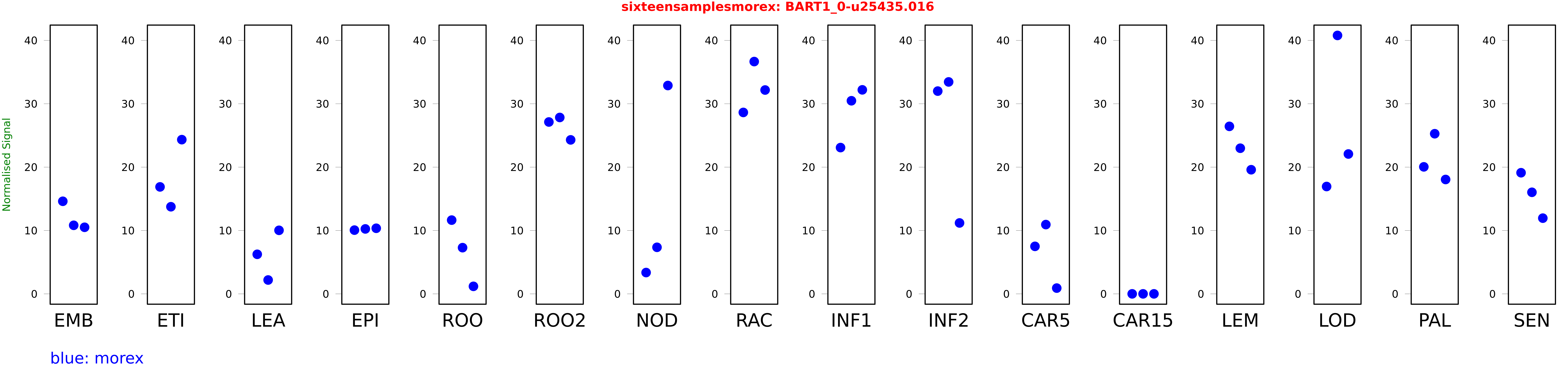
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 14.6124 | 10.8197 | 10.5066 |
| morex | ETI | 16.8819 | 13.75 | 24.3393 |
| morex | LEA | 6.24316 | 2.18582 | 10.0288 |
| morex | EPI | 10.0589 | 10.2494 | 10.3524 |
| morex | ROO | 11.6391 | 7.2903 | 1.1868 |
| morex | ROO2 | 27.128 | 27.8401 | 24.3038 |
| morex | NOD | 3.36363 | 7.34768 | 32.8841 |
| morex | RAC | 28.6347 | 36.6639 | 32.1735 |
| morex | INF1 | 23.0877 | 30.4778 | 32.2004 |
| morex | INF2 | 32.0032 | 33.4511 | 11.1908 |
| morex | CAR5 | 7.49837 | 10.9339 | 0.909129 |
| morex | CAR15 | 0 | 0 | 0 |
| morex | LEM | 26.4366 | 22.9866 | 19.5909 |
| morex | LOD | 16.9434 | 40.7902 | 22.0744 |
| morex | PAL | 20.0457 | 25.2739 | 18.0517 |
| morex | SEN | 19.1178 | 16.0304 | 11.9483 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g72890.2 | +3 | 3e-51 | 182 | 99/118 (84%) | protein|transposon protein, putative, CACTA, En/Spm sub-class, expressed |
| TAIR PP10 | AT1G16610 @ ARAPORT AT1G16610.1 @ TAIR |
+3 | 4e-40 | 150 | 82/118 (69%) | Symbols: SR45, RNPS1 | arginine/serine-rich 45 | chr1:5675925-5678686 REVERSE LENGTH=414 |
| BRACH PP3 | Bradi2g61380.4.p | +3 | 4e-52 | 184 | 105/119 (88%) | pacid=32777636 transcript=Bradi2g61380.4 locus=Bradi2g61380 ID=Bradi2g61380.4.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (1693 bp)
>BART1_0-u25435.016 1693 150831_barley_pseudomolecules ATATCCCCCCCGACCAAAACCCTAGCTCGCGCTCCAGACCCCGCACGCCCGCCGCCGCCG CGGGGAGACGGACGACGATGGCGAAGCCCCGCCGCGGCCGCTCAGGCTCGCGGTCCTCGT CGGGCTCCTCCTCCCGCTCCGCCTCCTCGGGCTCCTCCCGCTCGCGCTCGCGCTCCCGCT CCCGCTCCCTCTCCTCCTCCTCGTCCTCGCCCGCCCGCACCCGGGGCCGCTCCCCTCCCG CCGCCAAGCGCAGTTCACCTGGAGCAAAGAAAGGGCGATCACCTTCACCCCCTCTCAAAA AAGGGTCACCATCAAGGAAAGGCCGCTCAGCATCTCCCCCACCGAAGAAAGCTTCACCTC CTAGGAAAGCATCTCCCCCTCCTCAGTCGGTTGTTCTACACGTTGATCATCTATCCAGAA ATGTGAATGAGGCTCATTTGAAGGAGATATTTGGAAGTTATGGCGAAGTGGTGAATGTGG AGCTATCAATGGACAAATTTGTTAATCTCCCTCGTGGGTATGGATACATCGAGTTCAAGA AGAGGACTGATGCTGAGAAGGCTCTTCTTTACATGGATGGTGGGCAAATTGATGGAAATG TTGTCAAGCTGAAGTTCACACTGCCACCACGCCAAAAGGCTTCTTCACCTTTAAAGCCAC CTCCTCCTCCCCCCAGAAGAGATGCTCCTCAGAATGATAAAGTTGCTATTGGTGCTGAAA AGGATGCGCAGCAGCGCCCTGGGGGATCAGCATCTCCACGAAAGAAGCCACCTTCTCCTC CGCGAAAGAGGTCTCCTCCAAATAGAAGGGCTGAGTCACCCCGGCGTCCACCCAATCCAT CTCCAAGGCGTCGTGCTGATTCTCCTATTCGTCGTCGACCAGATCTTTCCCCTGTTAGGC GTGGTGACAGAAGACCAGGATCTCCAATTAGAAGACGTTCCCCTTCACCTAGAAGGCATA GGTCACCAATGCGCTTATCACCTCGCAGACGCTCCCCGGGGCCTCCTAGGCGCTCACCTG GGCCTCCCAGACGGCGGTCACCACCACCAAGGAGATTGAGGAGTCCTCCAAGAAGGCCAC CACCACCACCTCCCCGTCGTCACAGTCGCTCCCCTCCTCCTCGCCGCCCTCTTCACTCCC GCTCTAGATCGATTTCTCCTCGCAGGGGGCGAGGACCACCATTGAGGCGTGGAAGGTCGG ATTCATCATATTCTGCATCACCCAGTCCTCCAAGAAAGGGACCAAGAAGGGTGTCAAGGA GCCGCAGCCCTAGAAGGCCTCCCAGAGGAAGGAGCGGATCTAGCGACAGCGGGAGCAGCA GCTCTTCCCCAACCCCAAGGCGCAGGTAGAAAGAAAGAAACTCGTAACGTGTGTTGTTCC TGGCAGGTAAAAAAACTTTCTCTTCTCTCTCGACTGTCACCATGGATTGCGTCTGCATGC ATGTATCTTGCTGTGAGCTCAAGTCAAGAGGCGATCGATGTGAGGGGCGGGACGATGCGA TGAGGTGCTCGGCATGCCTCCCTGGCATTTGTACTCACAAGACAGACTGTATGATGACGA CTTGTGTTGATGATTTGCCTGTACTGGAACTATTTTTTTATAATTCTGCGGTTGTTTGTC AACTACAAATCTGGCTAGTATTATTATCCTGTTGCCACTCTTGAGAATTAATGAGTTGTT TACTAGTTGATTG
Protein Sequence (423 aa)
>BART1_0-u25435.016 423 150831_barley_pseudomolecules MAKPRRGRSGSRSSSGSSSRSASSGSSRSRSRSRSRSLSSSSSSPARTRGRSPPAAKRSS PGAKKGRSPSPPLKKGSPSRKGRSASPPPKKASPPRKASPPPQSVVLHVDHLSRNVNEAH LKEIFGSYGEVVNVELSMDKFVNLPRGYGYIEFKKRTDAEKALLYMDGGQIDGNVVKLKF TLPPRQKASSPLKPPPPPPRRDAPQNDKVAIGAEKDAQQRPGGSASPRKKPPSPPRKRSP PNRRAESPRRPPNPSPRRRADSPIRRRPDLSPVRRGDRRPGSPIRRRSPSPRRHRSPMRL SPRRRSPGPPRRSPGPPRRRSPPPRRLRSPPRRPPPPPPRRHSRSPPPRRPLHSRSRSIS PRRGRGPPLRRGRSDSSYSASPSPPRKGPRRVSRSRSPRRPPRGRSGSSDSGSSSSSPTP RRR
