- Transcript BART1_0-u25331.005
- Transcript IDBART1_0-u25331.005
- Gene IDBART1_0-u25331
Exon Structure of BART1_0-u25331.005
| Chromosome | Exon | Start | Stop | Direction |
|---|---|---|---|---|
| chr3H | 1 | 679779870 | 679779657 | + |
| chr3H | 2 | 679780077 | 679779949 | + |
| chr3H | 3 | 679780262 | 679780159 | + |
| chr3H | 4 | 679780772 | 679780595 | + |
| chr3H | 5 | 679781035 | 679780884 | + |
| chr3H | 6 | 679782367 | 679781761 | + |
| chr3H | 7 | 679783932 | 679783777 | + |
| chr3H | 8 | 679784080 | 679784000 | + |
| chr3H | 9 | 679784262 | 679784187 | + |
| chr3H | 10 | 679784610 | 679784345 | + |
| chr3H | 11 | 679785333 | 679785100 | + |
| chr3H | 12 | 679786251 | 679785445 | + |
| chr3H | 13 | 679786878 | 679786479 | + |
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
Salmon TPM Values: sixteensamplesmorex
These TPM values were generated by using the RNA-seq data from a 16-tissue experiment in Morex (published here) were calculated using Salmon (version Salmon-0.8.2) using BaRTv1.0-QUASI as the reference transcript dataset. Click here for more information about the RNA-seq experiment and materials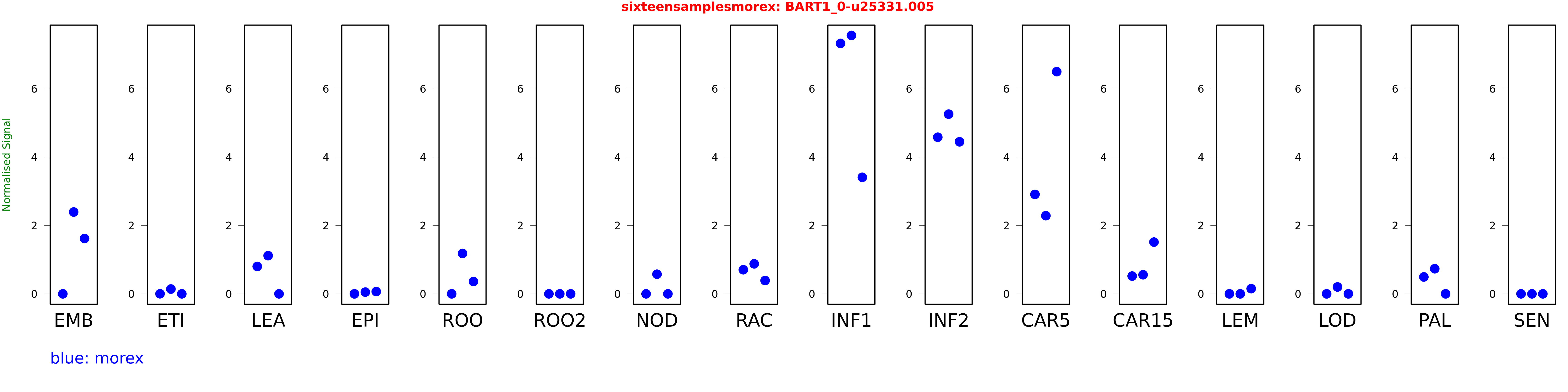
| Sample | Treatment | Rep 1 | Rep 2 | Rep 3 |
|---|---|---|---|---|
| morex | EMB | 0 | 2.39448 | 1.61913 |
| morex | ETI | 0.00228375 | 0.140788 | 0.000302018 |
| morex | LEA | 0.801209 | 1.11748 | 0.000000775742 |
| morex | EPI | 0 | 0.0512137 | 0.0663872 |
| morex | ROO | 0 | 1.18231 | 0.359964 |
| morex | ROO2 | 0 | 0 | 0 |
| morex | NOD | 0 | 0.574463 | 0 |
| morex | RAC | 0.705092 | 0.877868 | 0.390209 |
| morex | INF1 | 7.32825 | 7.56118 | 3.41019 |
| morex | INF2 | 4.58266 | 5.25806 | 4.44834 |
| morex | CAR5 | 2.90924 | 2.28677 | 6.50074 |
| morex | CAR15 | 0.520014 | 0.558937 | 1.51388 |
| morex | LEM | 0 | 0 | 0.151881 |
| morex | LOD | 0 | 0.201909 | 0 |
| morex | PAL | 0.494807 | 0.735039 | 0 |
| morex | SEN | 0 | 0 | 0 |
Homology to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Rice PP7 | LOC_Os01g68810.1 | +2 | 0.0 | 1395 | 770/1027 (75%) | protein|expressed protein |
| TAIR PP10 | AT1G24706 @ ARAPORT AT1G24706.2 @ TAIR |
+2 | 0.0 | 818 | 511/1022 (50%) | Symbols: THO2 | THO2 | chr1:8742210-8755250 FORWARD LENGTH=1823 |
| BRACH PP3 | Bradi2g58697.4.p | +2 | 0.0 | 1596 | 839/967 (87%) | pacid=32780758 transcript=Bradi2g58697.4 locus=Bradi2g58697 ID=Bradi2g58697.4.v3.1 annot-version=v3.1 |
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
grey: non-coding, green: Barley Morex IBSC 2017 Transcript CDS, red: Barley RTD exons
CDS Sequence (3404 bp)
>BART1_0-u25331.005 3404 150831_barley_pseudomolecules GCAATAGTTCTATATATCCTTCTAATCACTGGGGGTGCTTGTGTTTCCCCTTCCAGTATT GTTAGTCTTTAATCTTCATCTTTATGCTTCTATTCTGAACCAACAAACTTTTAACAGCTG CAAGAAGCATCTTTCCATGGAGCTTAAATGCCTTTTCCAATACATAGTTAATCAACTGAA GAAAGGATTGGGCACTGAGCTTGTTGTACTTGAGGAACTCATTCAGCAAATGGCAAATGT ACAGTACACTGAGAACATGACTGATGAACAGGTTGATGCTATGGCAGGAAGTGAAACATT GCGGCTTCAATCTTCGCTGTTTGGATCAACACGGAACTATAAGGTCCTGAATAAGTCTAC TAACAAATTGCGGGATTCTTTGCTTCCAAAAGATGAACCTAAACTGGCAATTCCTCTACT ACTGCTCGTTGCCCAGCACAGATCCAAGATTATAATAAATGCAGATGCCACATATATCAA AATGGTTAGCGAGCAGTTTGATAGATGCCATGGAATACTACTTCAGTATGCCGAGTTTCT TTCAAGTGCTGTAACTTCATCAACCTATGTTCAGCTAGTACCTCCGCTGGAAGATCTCGT TTATAAATATCATATCGAGCCAGATATTGCTTTTCTTATATACCGCCCTGTAATGAGGCT TTTCAAAAGCGCTAATGGTGGCGAGGCCTGTTGGCCTCTGGATGACAATGAAGAAGGGGA GTCTGTATCATGTGATGAAATGATCTTGCATGGTGATTCATCTCAGAAGTTAATCATGTG GTCAGATCTTCTCAACACAATCCGAACAATTTTGCCAGCAAAAGCCTGGAACGGCCTTTC ACCCGAATTGTATGCTACTTTCTGGGGTTTAACACTGTATGATCTTCATTTTCCAAAAGA TCGTTATGATGCAGAAATCAAGAAGCTGCATGAAAATCTCAAACAACTTGAAGATAACTC AGATAACTCTAGCATTGCAATATCAAGACGTAAGAAGGATAAGGAGAGAATCCAAGATTT GCTGGACAAATTGAACAATGAATCTGACAAGCATCAGCACCATGTTCTATCAGTGCTCCA AAGGCTAACCCGTGAAAAGGATAAATGGCTGAGTTCCAGTCCGGACGCACTGAAGATCAA CATGGAATTTCTCCAGCGTTGCATATATCCACGCTGTGTTCTTAGCATGCAGGATGCTGT ATATTGTGCTACATTCGTCCAGATGATGCATTCACTTGGGACTCCATTTTTCAACACGGT GAATCATATAGATGTTTTCATCTGCAAGACACTACAGCCAATGATCTGTTGTTGTACAGA ATATGAGGCTGGCAGGCTGGGAAGGTTTCTTCATGAGACCCTGAAGATGGCCTACCATTG GAAGGTTCATTGGAAATGGAGTGGAAGAATTACCAAGGTGCTTAATCAATGTATGGAGTC AAAGGAATACATGGAAATCAGAAACGCCCTTATTGTGCTCACAAAGATAACTAGTATTTT TCCTGTGATGCGAAAAAGCGGTATCAACATCGAGAAAAGGGTTGCGAAGCTAAAGGGGGA TGAGAGGGAAGATCTGAAAGTACTGGCCACTGGTGTAGCTGCTGCTCTGGCCGCTCGCAA GAGTTCATGGGTGTCTGAAGAAGAGTTTGGCATGGGTCACCTTGATCTGAAGCCAGTGCT TGCAAAACCTATTGCTGGAAATCAGTATGCAGATCCCTCGACAGCAAAAGATCATAGCGT TCGTGCAAAATCTGTAGAGGGTAGGCATGAAAGGTCAGAAAATGCAATGAAGCCTGATGC TCAACATAAGAAGAATGCCTCAACTACAAACGGGTCTGATAGTCAAATACCATCTTCCTC TGCACAAGGAAAAGGTTCAGGGATTGCACGCGGTGCAGATGAACCTCCAAAACTTTTGTC CGATGATGGAGTTAAAGTCTTGAAGCCTACTGCAGAACCCGAGACAAGAGCGTCACAAAA GCGTGCTGTACACAATGCTGCAAAGGTATCTAAGCATGATGTGGTGAAGGAAGATGCAAA ACCTGGGAGATCTACTAGCAGGGGTCTTAATCAACAAGCCTCTGCTATTCCTGTTGACAA AGAAGTATTGTCTCAGGCAGCTGATGGTGTGCTGGATACTAATCCCACTAGTCCGTTGGT TGGCACAAATGGTAATGTACATCCGGCACCACGAAAGCAAAAAAGATCTGTCGCCGTTGA AGAACAAGAGAGGACAGGTAAACGAAGGAAAGGAGAGATTGAAGGCCGGGATGGTGACTT GGCAGAACACCATACGGACAAAGAAAAAAAATTGGACCCGCGATCAGTAGATAAATTTCG TTCTGTGGATCATGAGAGAGGTGCTAGTGAGGAACAAAACCTAATTCGGACAGAGAAATT AAAAGAAAAGTTCGATGAGAAATATGAAAGAGATCATAGAGAAAAAGCAGATCGGTCTGA GAGGCGCCGTGGAGAAGACGTGGTTGAGAGACCAACAGACAGATCATTGGAAAGGAGAGA GCGTTCTATTGAAAAGATGCAAGAGAGAGTTCCTGAAAAAGGAAGAGAAGACAGGAATAA GGAGGAGAGGAACAAAATTAAGCATGAACCCATAGACCGAGCACATACTATTAAGAATGA ACCCATAGACCGAGCACATACTATTAAGCATGAACCCATAGACCGAGCACATACTATTAA GCATGAACCCATAGACCGAGCACATACTTCTGACGAACGCTTCCGAGGGCAAAGCTTGCC ACCACCTCCACCTCTTCCAACTAGTTTTGTACCCCAATCAGTCGCTGCTAACCGAAGAGA TGAAGACATTGACCGAAGGGGGAGCAGCACCAGGCATACACAGCGGTTGTCTCCCAGGCG TGATGAGAAAGAAAGGTGGCACTTGGAGGAGAATGCTCCGCTGTTGCAGGATGATGGGAA GCACAGGAGAGAGGAAGATCTCCGGGATAGAAAGCGTGAAGATAGGGATGTTTCATCAAG CAAGACGACAGGGATAGGGACAAAGGTAATACTGTGAAGGAAGATAGTGATCCTAATAGT GCATCCAAACGTCGAAAGATTAAAAGAGAACAGTCTGCTTTGGAGGCTGGGGAATACGCG CCTTCTGCTCCACAGCCTCCGAGCGTTGGACCTGGTAACTCACAGTATGAAATACGAGAG AGAGAAAGAAAGGGTGCTATTTCACAGCATCGTCCATCACATGCTGATGACCTTCCTAGG ATGCATGCCAAGGATTCAACAAGCAAGACAAGTCGTAGAGAGGCTGACCAGTAAGTTTAG GAAACAATGATCTCATTGAACCATGCATTACAACAGTTTCCACTTTGCAGTATGTTCTTT ATACCTTATTAGCTGTCATGTGGAACACACAATCTGTTTATGGG
Protein Sequence (966 aa)
>BART1_0-u25331.005 966 150831_barley_pseudomolecules MELKCLFQYIVNQLKKGLGTELVVLEELIQQMANVQYTENMTDEQVDAMAGSETLRLQSS LFGSTRNYKVLNKSTNKLRDSLLPKDEPKLAIPLLLLVAQHRSKIIINADATYIKMVSEQ FDRCHGILLQYAEFLSSAVTSSTYVQLVPPLEDLVYKYHIEPDIAFLIYRPVMRLFKSAN GGEACWPLDDNEEGESVSCDEMILHGDSSQKLIMWSDLLNTIRTILPAKAWNGLSPELYA TFWGLTLYDLHFPKDRYDAEIKKLHENLKQLEDNSDNSSIAISRRKKDKERIQDLLDKLN NESDKHQHHVLSVLQRLTREKDKWLSSSPDALKINMEFLQRCIYPRCVLSMQDAVYCATF VQMMHSLGTPFFNTVNHIDVFICKTLQPMICCCTEYEAGRLGRFLHETLKMAYHWKVHWK WSGRITKVLNQCMESKEYMEIRNALIVLTKITSIFPVMRKSGINIEKRVAKLKGDEREDL KVLATGVAAALAARKSSWVSEEEFGMGHLDLKPVLAKPIAGNQYADPSTAKDHSVRAKSV EGRHERSENAMKPDAQHKKNASTTNGSDSQIPSSSAQGKGSGIARGADEPPKLLSDDGVK VLKPTAEPETRASQKRAVHNAAKVSKHDVVKEDAKPGRSTSRGLNQQASAIPVDKEVLSQ AADGVLDTNPTSPLVGTNGNVHPAPRKQKRSVAVEEQERTGKRRKGEIEGRDGDLAEHHT DKEKKLDPRSVDKFRSVDHERGASEEQNLIRTEKLKEKFDEKYERDHREKADRSERRRGE DVVERPTDRSLERRERSIEKMQERVPEKGREDRNKEERNKIKHEPIDRAHTIKNEPIDRA HTIKHEPIDRAHTIKHEPIDRAHTSDERFRGQSLPPPPPLPTSFVPQSVAANRRDEDIDR RGSSTRHTQRLSPRRDEKERWHLEENAPLLQDDGKHRREEDLRDRKREDRDVSSSKTTGI GTKVIL
